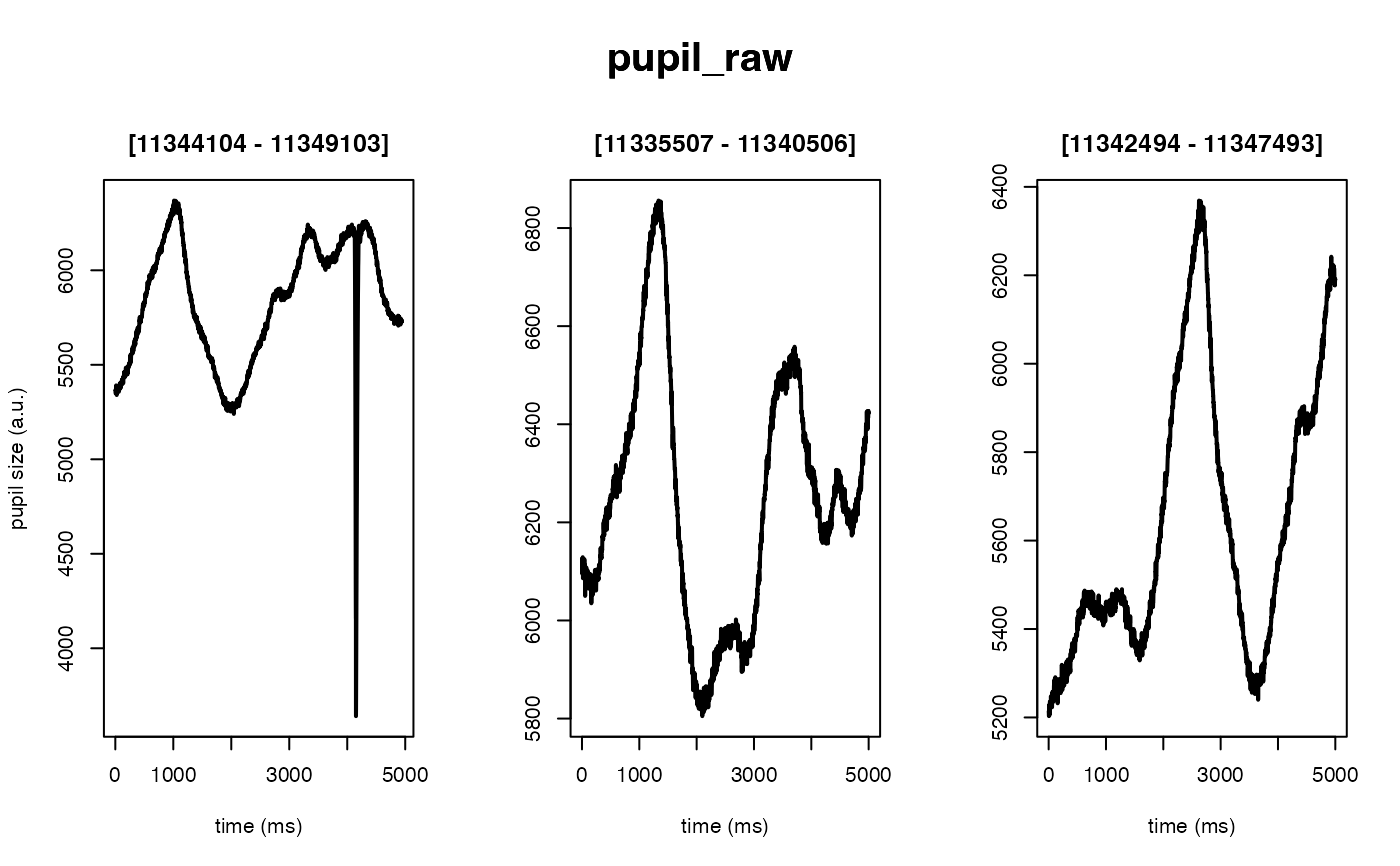
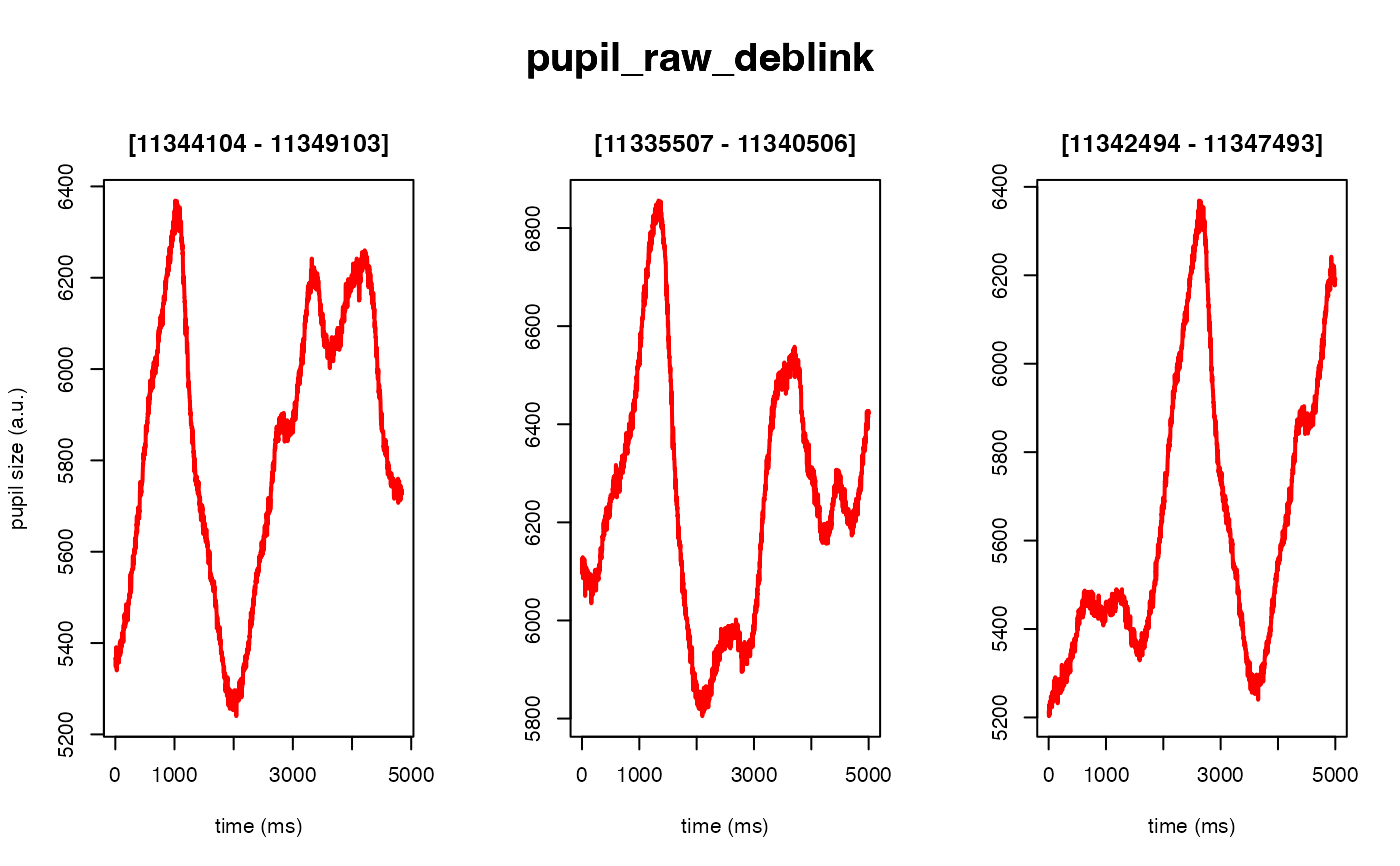
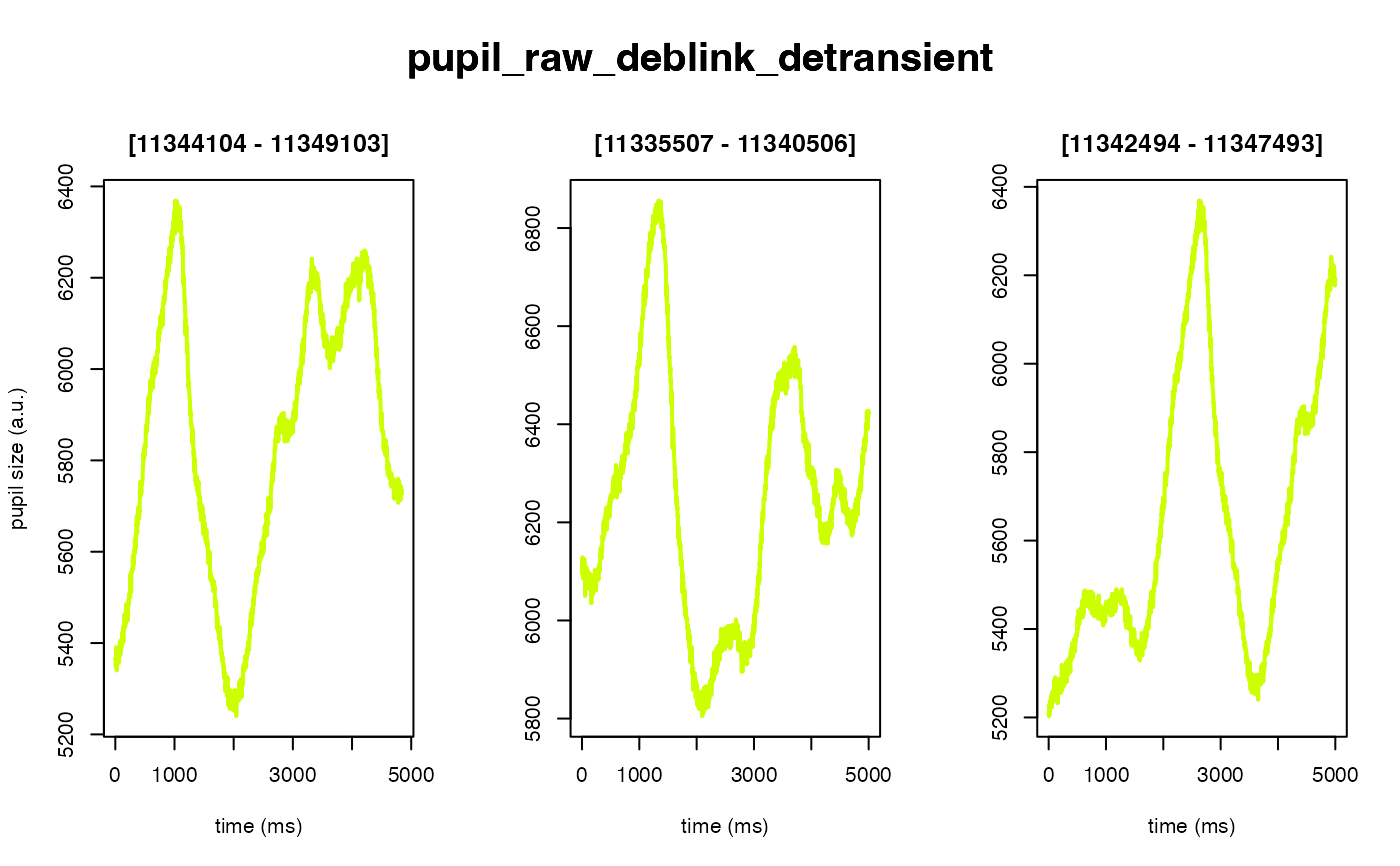
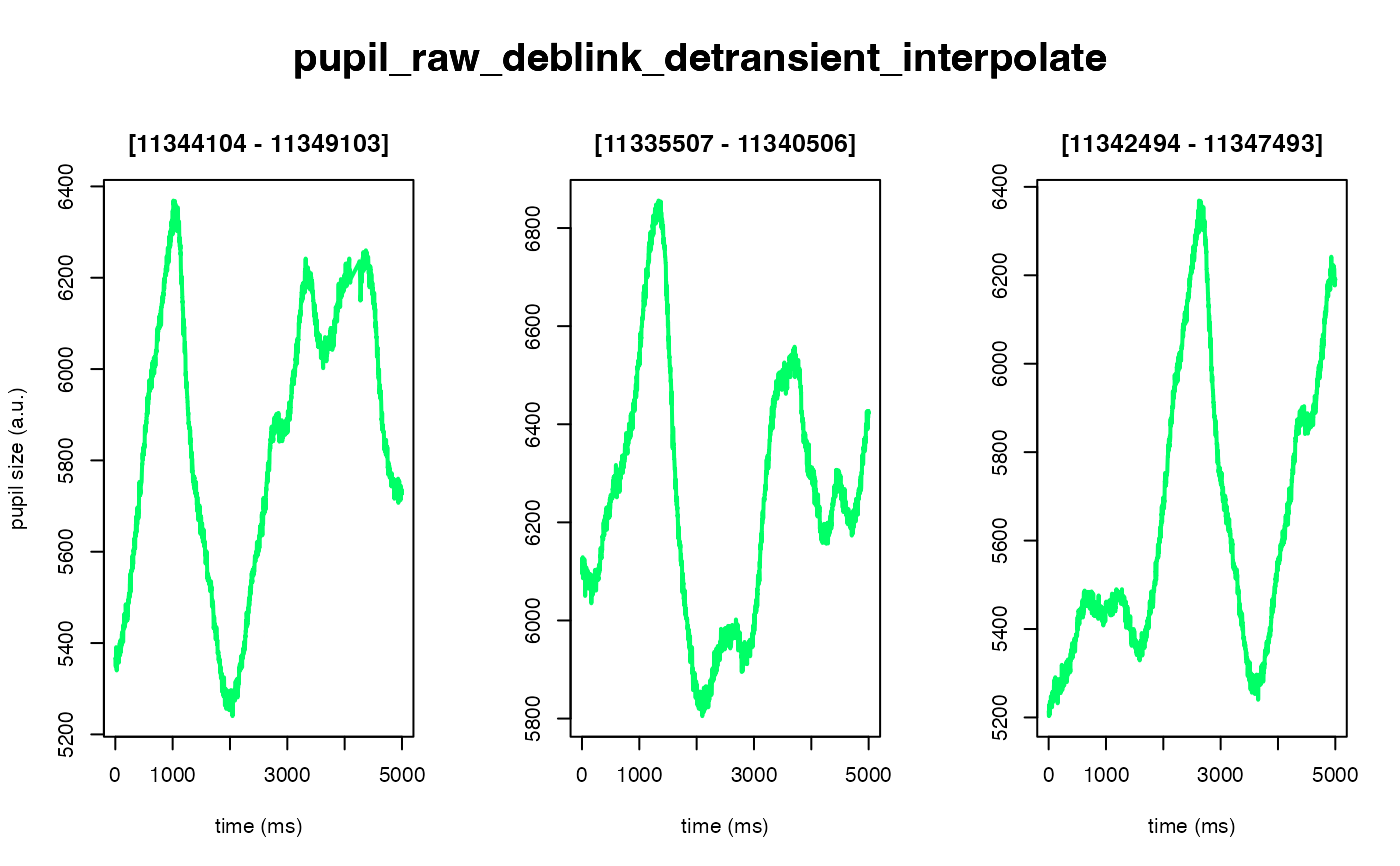
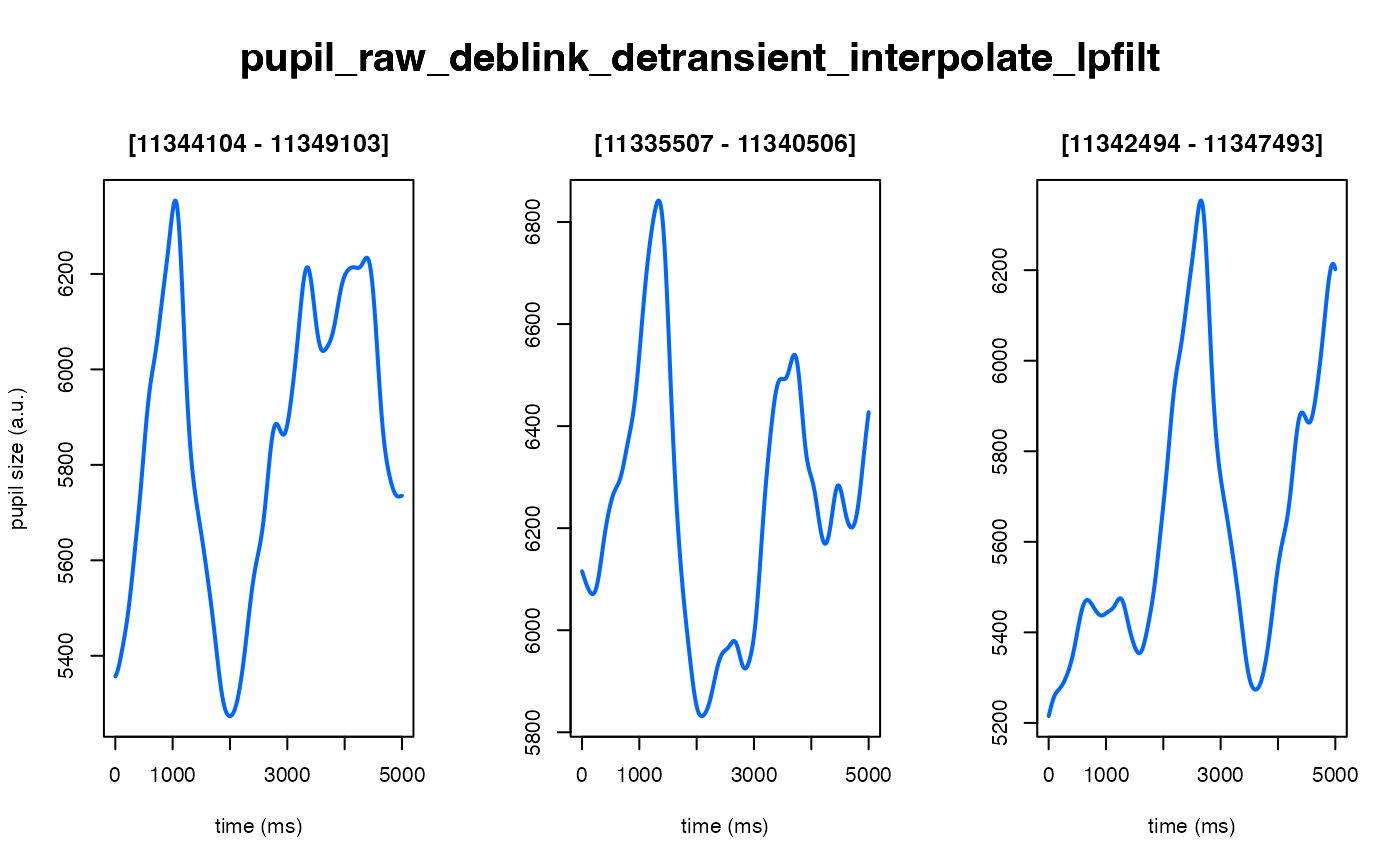
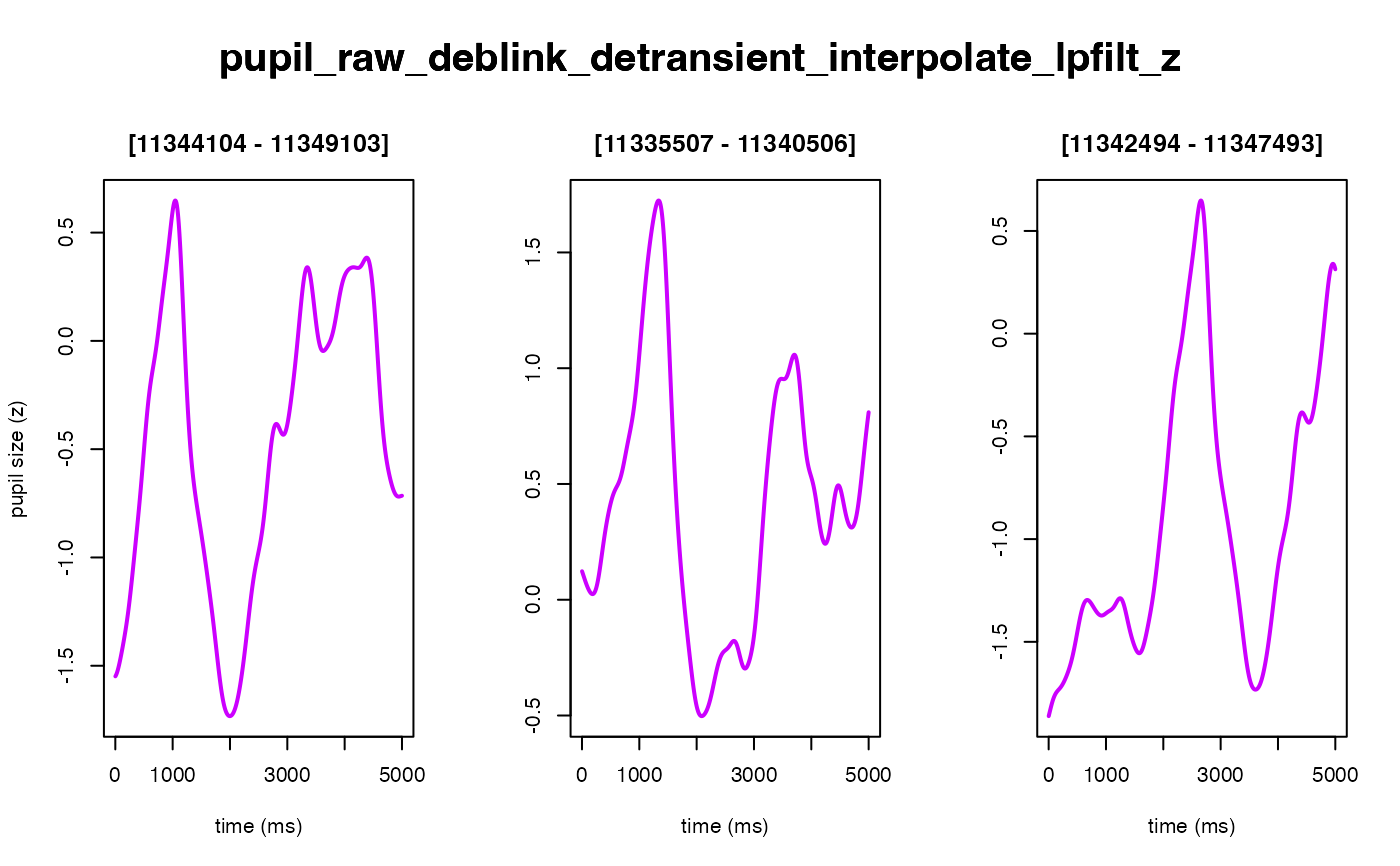
The intended use of this method is to scale the arbitrary units of the pupil
size timeseries to have a mean of 0 and a standard deviation of 1. This
is accomplished by mean centering the data points and then dividing them by
their standard deviation (i.e., z-scoring the data, similar to
base::scale()). Opting to z-score your pupil data helps with trial-level
and between-subjects analyses where arbitrary units of pupil size recorded by
the tracker do not scale across participants, and therefore make analyses
that depend on data from more than one participant difficult to interpret.
Arguments
- eyeris
An object of class
eyerisdervived fromload().
Details
This function is automatically called by glassbox() by default. Use
glassbox(zscore = FALSE) to disable this step as needed.
Users should prefer using glassbox() rather than invoking this function
directly unless they have a specific reason to customize the pipeline
manually.
In general, it is common to z-score pupil data within any given participant, and furthermore, z-score that participant's data as a function of block number (for tasks/experiments where participants complete more than one block of trials) to account for potential time-on-task effects across task/experiment blocks.
As such, if you use the eyeris package as intended, you should NOT need
to specify any groups for the participant/block-level situations described
above. This is because eyeris is designed to preprocess a single block of
pupil data for a single participant, one at a time. Therefore, when you later
merge all of the preprocessed data from eyeris, each individual,
preprocessed block of data for each participant will have already been
independently scaled from the others.
Additionally, if you intend to compare mean z-scored pupil size across task conditions, such as that for memory successes vs. memory failures, then do NOT set your behavioral outcome (i.e., success/failure) variable as a grouping variable within your analysis. If you do, you will consequently obtain a mean pupil size of 0 and standard deviation of 1 within each group (since the scaled pupil size would be calculated on the timeseries from each outcome variable group, separately). Instead, you should compute the z-score on the entire pupil timeseries (before epoching the data), and then split and take the mean of the z-scored timeseries as a function of condition variable.
Note
This function is part of the glassbox() preprocessing pipeline and is not
intended for direct use in most cases. Use glassbox(zscore = TRUE).
Advanced users may call it directly if needed.
See also
glassbox() for the recommended way to run this step as
part of the full eyeris glassbox preprocessing pipeline.
Examples
demo_data <- eyelink_asc_demo_dataset()
demo_data |>
eyeris::glassbox(zscore = TRUE) |> # set to FALSE to skip (not recommended)
plot(seed = 0)
#> ✔ [ OK ] - Running eyeris::load_asc()
#> ✔ [ OK ] - Running eyeris::deblink()
#> ✔ [ OK ] - Running eyeris::detransient()
#> ✔ [ OK ] - Running eyeris::interpolate()
#> ✔ [ OK ] - Running eyeris::lpfilt()
 #> ! [ SKIP ] - Skipping eyeris::detrend()
#> ✔ [ OK ] - Running eyeris::zscore()
#> ! Plotting block 1 from possible blocks: 1
#> ! [ SKIP ] - Skipping eyeris::detrend()
#> ✔ [ OK ] - Running eyeris::zscore()
#> ! Plotting block 1 from possible blocks: 1