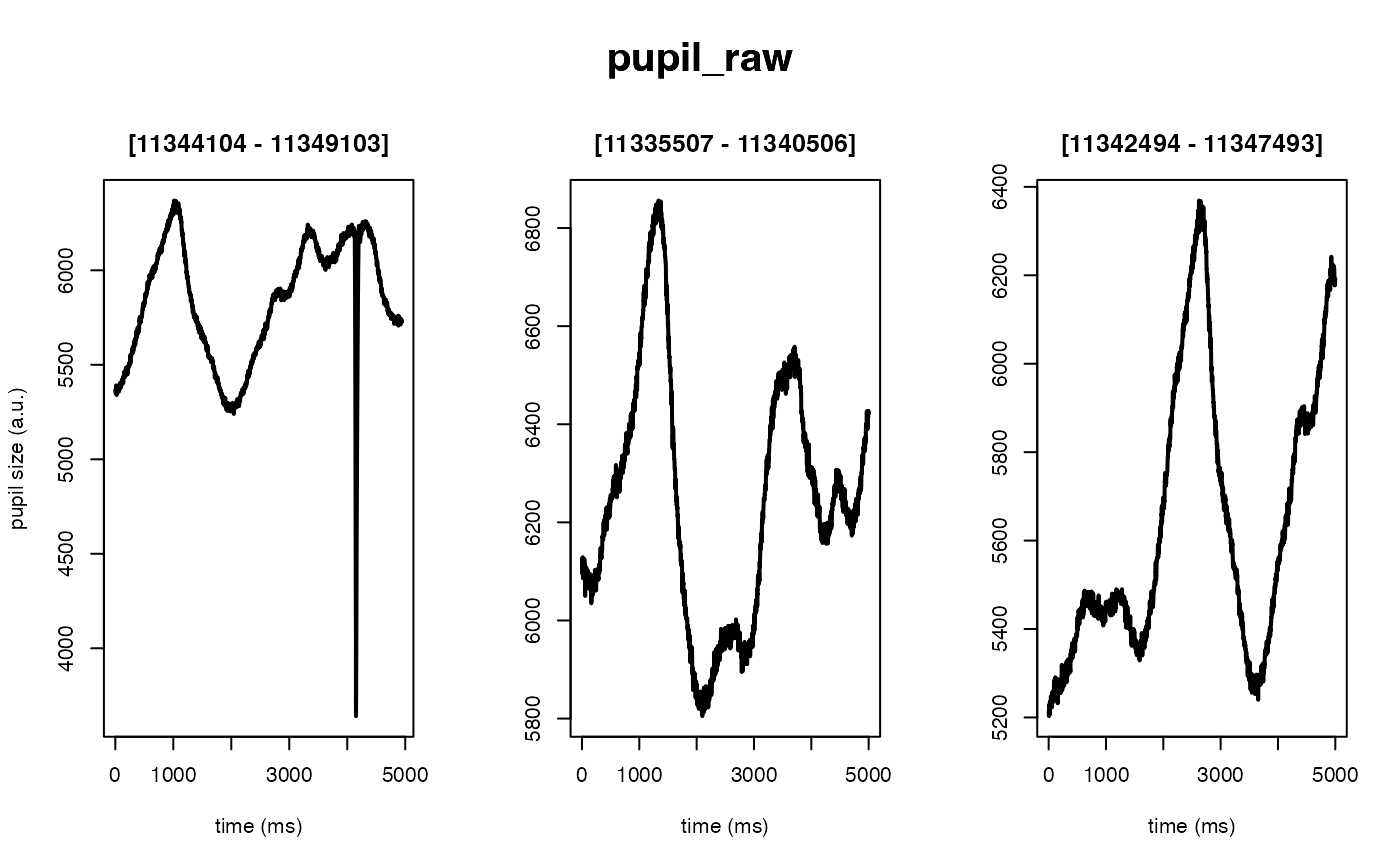
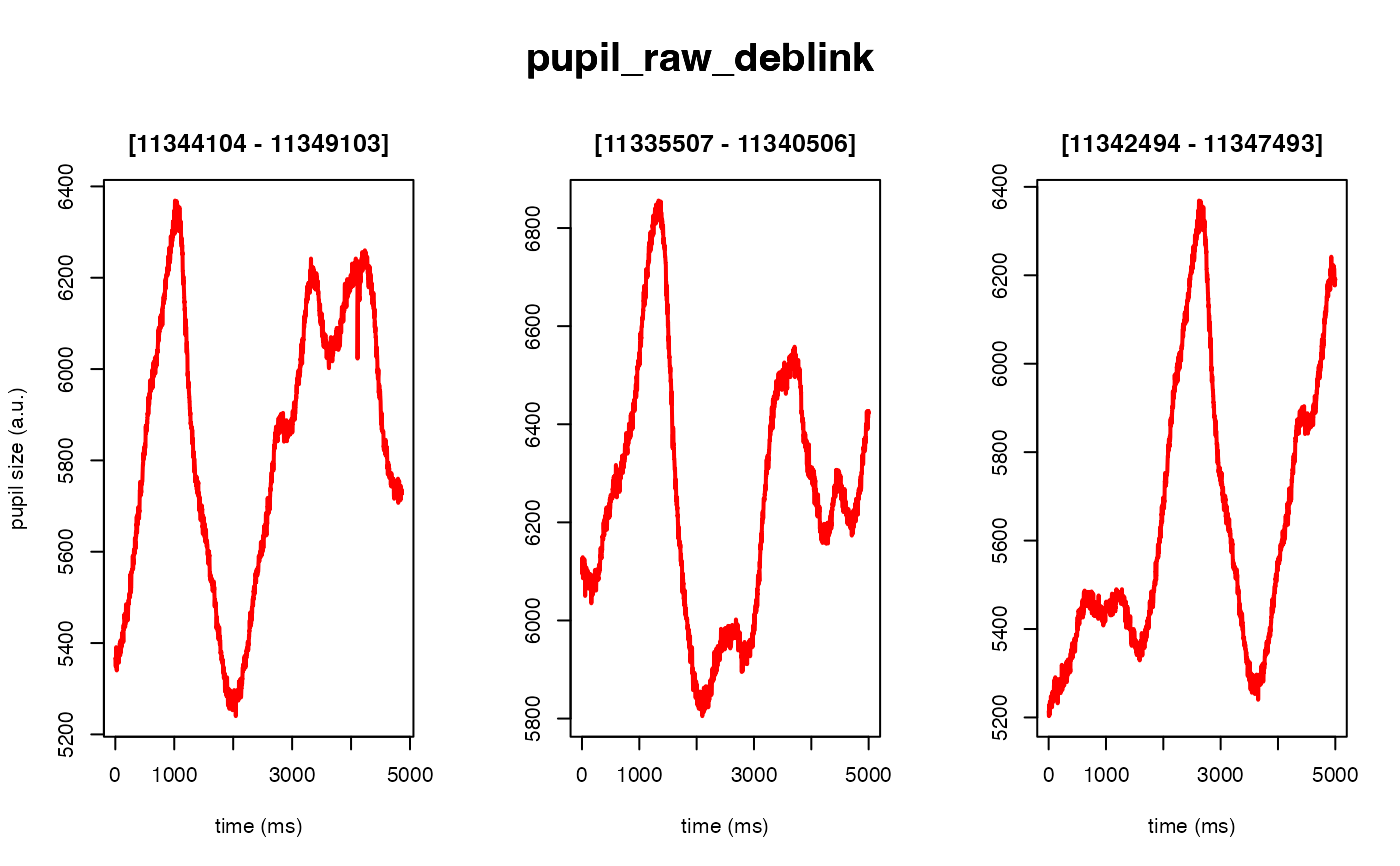
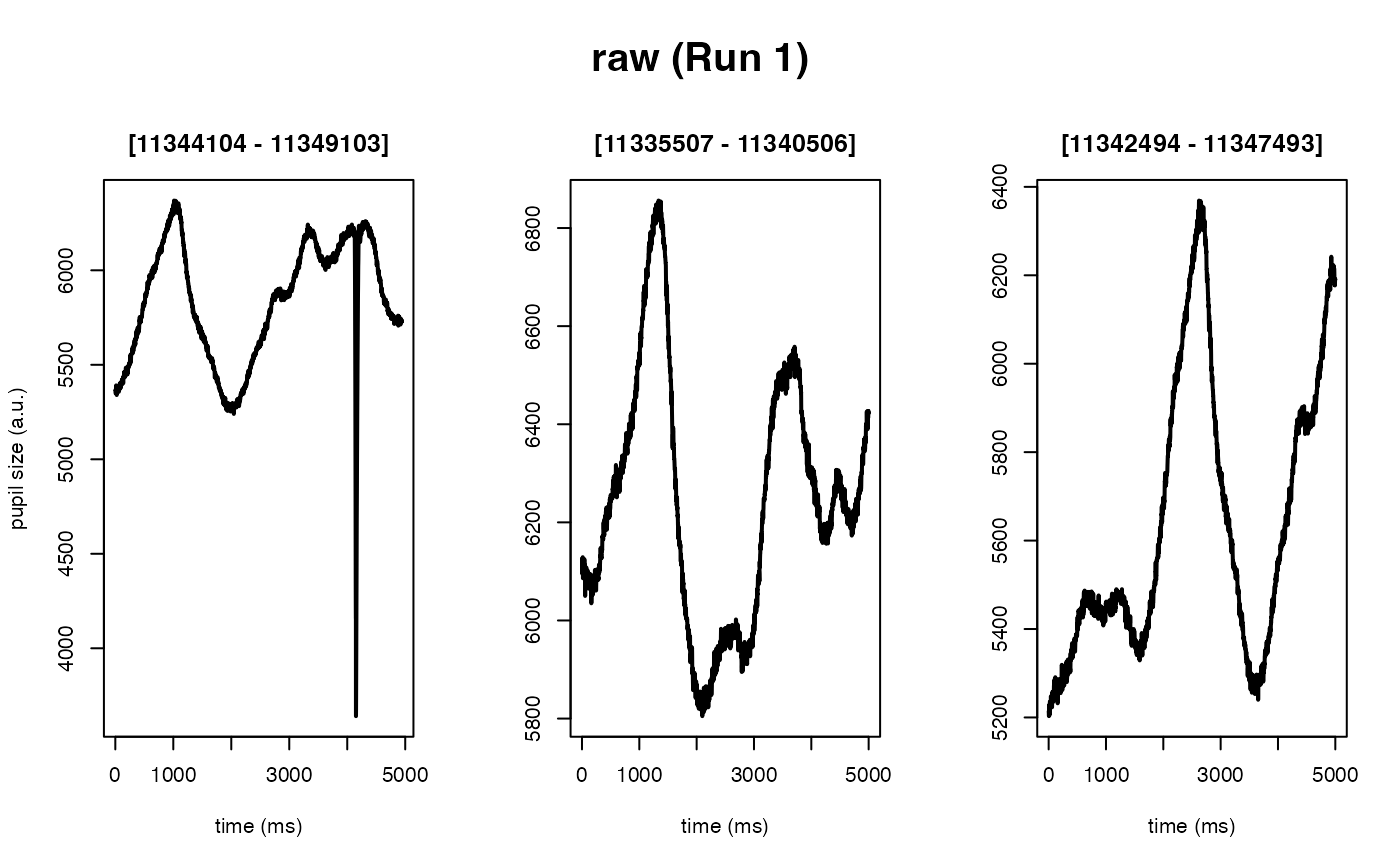
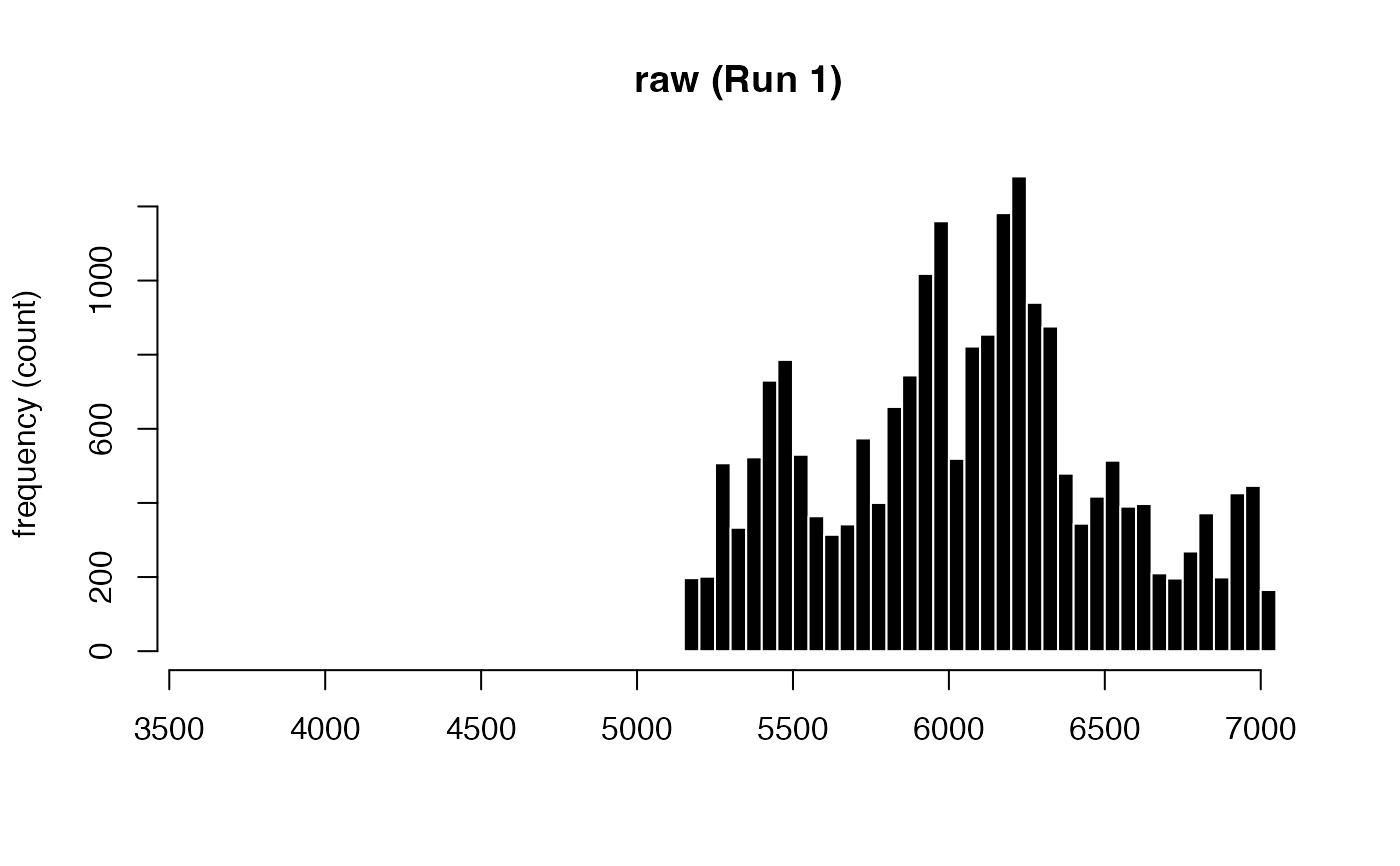
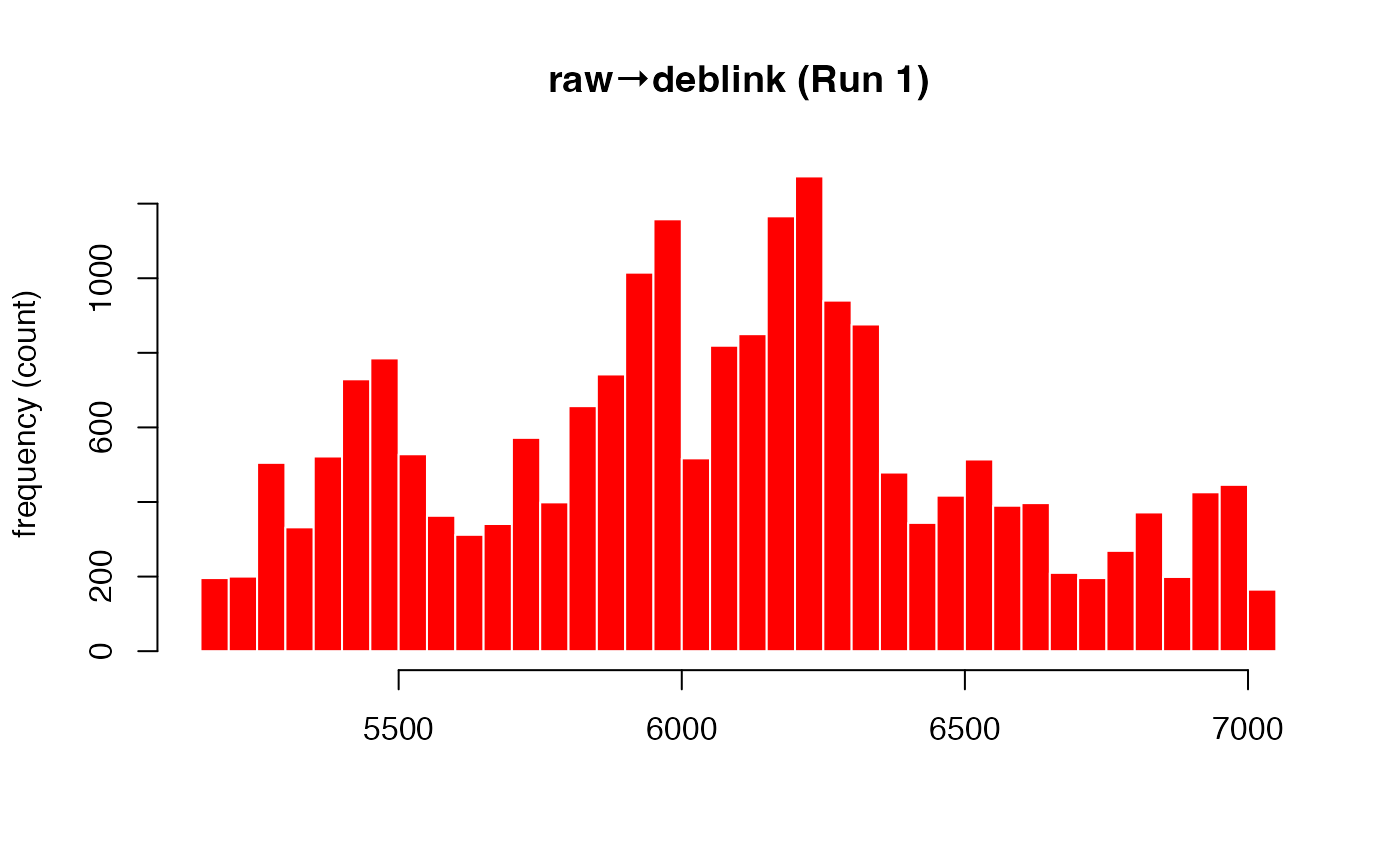
Deblinking (a.k.a. NA-padding) of time series data. The intended use of this method is to remove blink-related artifacts surrounding periods of missing data. For instance, when an individual blinks, there are usually rapid decreases followed by increases in pupil size, with a chunk of data missing in-between these 'spike'-looking events. The deblinking procedure here will NA-pad each missing data point by your specified number of ms.
Arguments
- eyeris
An object of class
eyerisderived fromload_asc()- extend
Either a single number indicating the number of milliseconds to pad forward/backward around each missing sample, or, a vector of length two indicating different numbers of milliseconds pad forward/backward around each missing sample, in the format
c(backward, forward)- call_info
A list of call information and parameters. If not provided, it will be generated from the function call
Details
This function is automatically called by glassbox() by default. If needed,
customize the parameters for deblink by providing a parameter list. Use
glassbox(deblink = FALSE) to disable this step as needed.
Users should prefer using glassbox() rather than invoking this function
directly unless they have a specific reason to customize the pipeline
manually.
Note
This function is part of the glassbox() preprocessing pipeline and is not
intended for direct use in most cases. Provide parameters via
deblink = list(...).
Advanced users may call it directly if needed.
See also
glassbox() for the recommended way to run this step as
part of the full eyeris glassbox preprocessing pipeline
Examples
demo_data <- eyelink_asc_demo_dataset()
# 50 ms in both directions (the default)
demo_data |>
eyeris::glassbox(deblink = list(extend = 50)) |>
plot(seed = 0)
#> ✔ [ OK ] - Running eyeris::load_asc()
#> ℹ [ INFO ] - Processing block: block_1
#> ✔ [ OK ] - Running eyeris::deblink() for block_1
#> ✔ [ OK ] - Running eyeris::detransient() for block_1
#> ✔ [ OK ] - Running eyeris::interpolate() for block_1
#> ✔ [ OK ] - Running eyeris::lpfilt() for block_1
 #> ! [ SKIP ] - Skipping eyeris::downsample() for block_1
#> ! [ SKIP ] - Skipping eyeris::bin() for block_1
#> ! [ SKIP ] - Skipping eyeris::detrend() for block_1
#> ✔ [ OK ] - Running eyeris::zscore() for block_1
#>
#> Block processing summary:
#> block_1: OK (steps: 6, latest: pupil_raw_deblink_detransient_interpolate_lpfilt_z)
#>
#> ✔ [ OK ] - Running eyeris::summarize_confounds()
#> ! [ INFO ] - Plotting block 1 from possible blocks: 1
#> ℹ [ INFO ] - Plotting with sampling rate: 1000 Hz
#> ! [ SKIP ] - Skipping eyeris::downsample() for block_1
#> ! [ SKIP ] - Skipping eyeris::bin() for block_1
#> ! [ SKIP ] - Skipping eyeris::detrend() for block_1
#> ✔ [ OK ] - Running eyeris::zscore() for block_1
#>
#> Block processing summary:
#> block_1: OK (steps: 6, latest: pupil_raw_deblink_detransient_interpolate_lpfilt_z)
#>
#> ✔ [ OK ] - Running eyeris::summarize_confounds()
#> ! [ INFO ] - Plotting block 1 from possible blocks: 1
#> ℹ [ INFO ] - Plotting with sampling rate: 1000 Hz
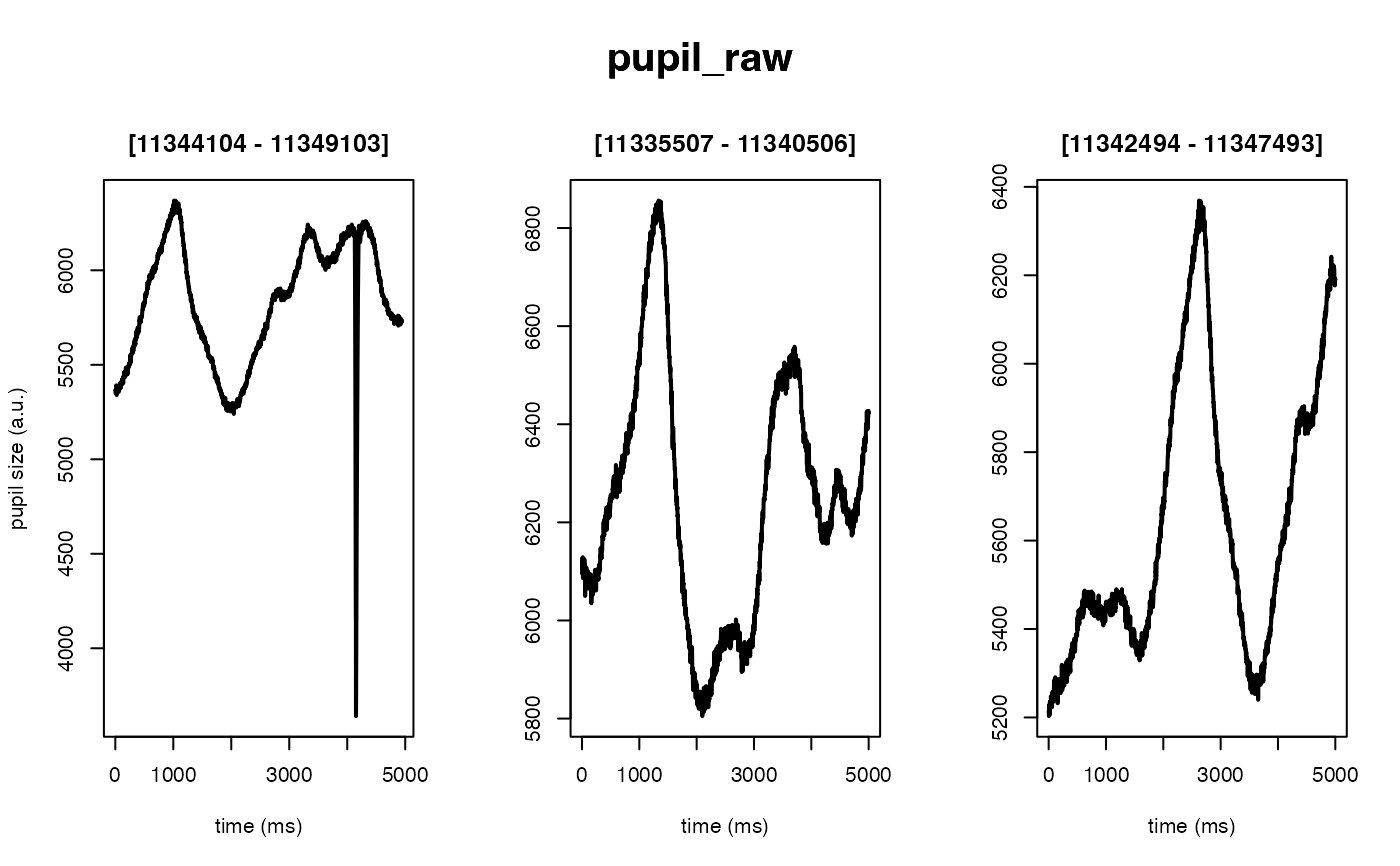
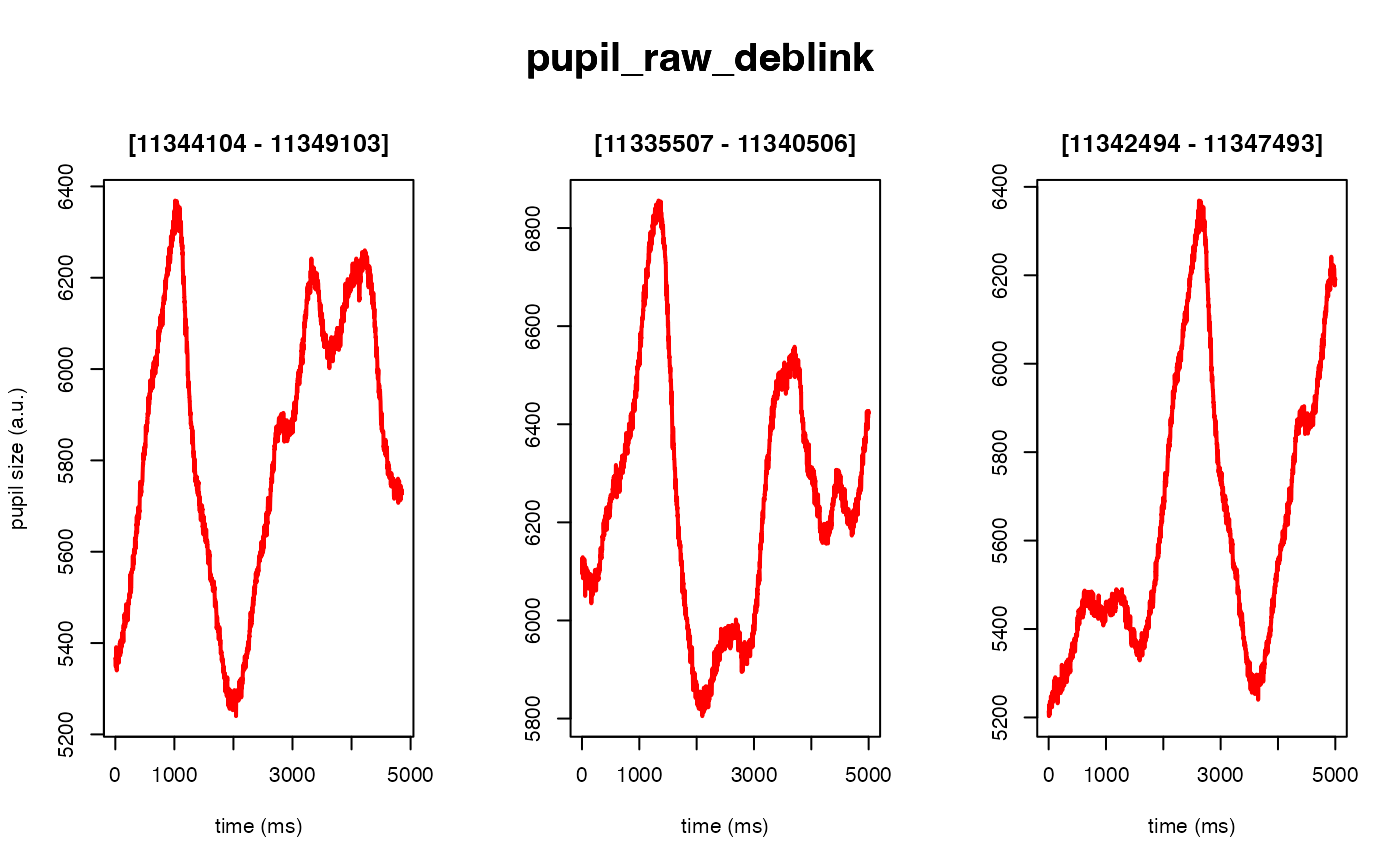
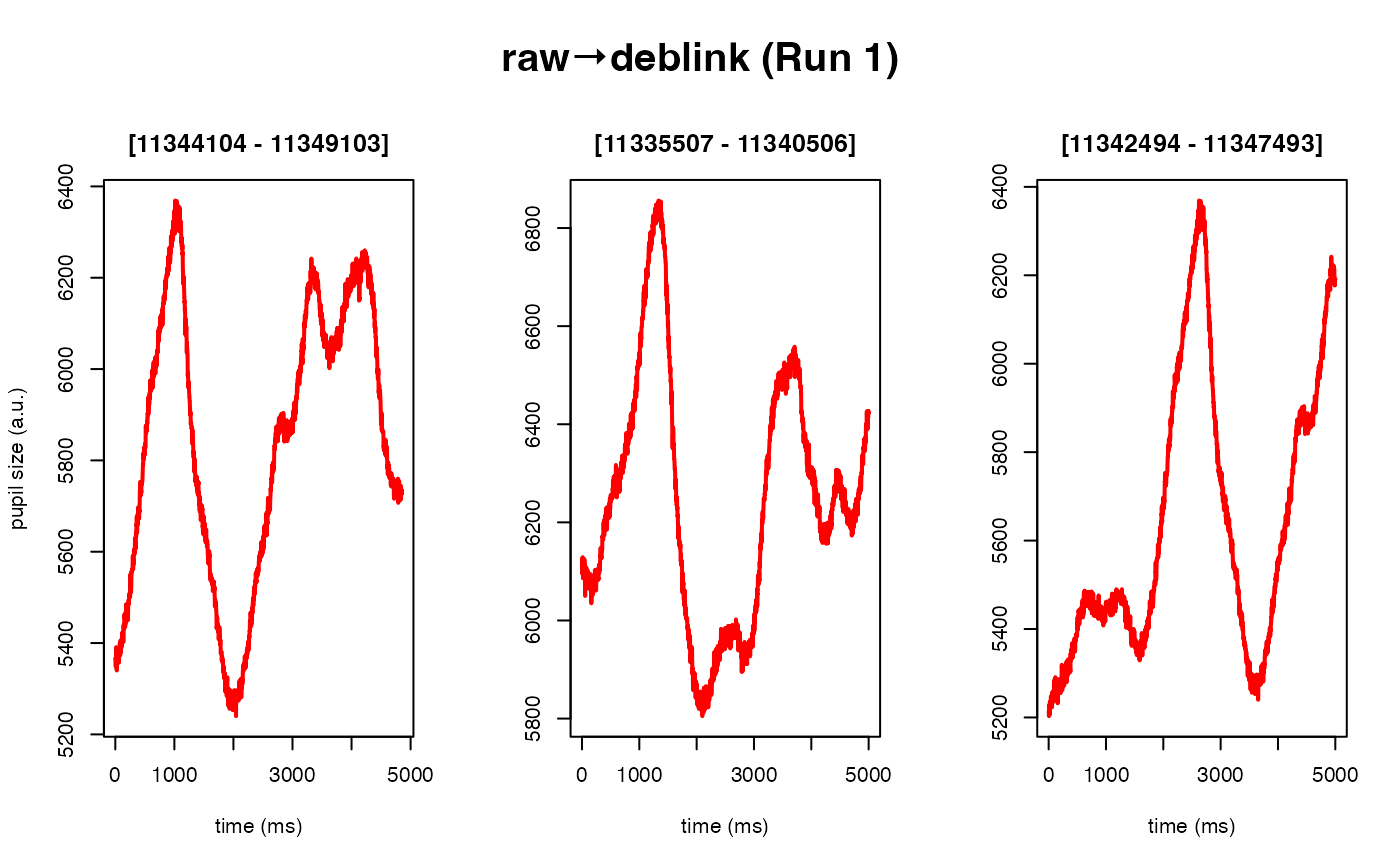 # 40 ms backward, 50 ms forward
demo_data |>
# set deblink to FALSE (instead of a list of params)
# to skip step (not recommended)
eyeris::glassbox(deblink = list(extend = c(40, 50))) |>
plot(seed = 0)
#> ✔ [ OK ] - Running eyeris::load_asc()
#> ℹ [ INFO ] - Processing block: block_1
#> ✔ [ OK ] - Running eyeris::deblink() for block_1
#> ✔ [ OK ] - Running eyeris::detransient() for block_1
#> ✔ [ OK ] - Running eyeris::interpolate() for block_1
#> ✔ [ OK ] - Running eyeris::lpfilt() for block_1
# 40 ms backward, 50 ms forward
demo_data |>
# set deblink to FALSE (instead of a list of params)
# to skip step (not recommended)
eyeris::glassbox(deblink = list(extend = c(40, 50))) |>
plot(seed = 0)
#> ✔ [ OK ] - Running eyeris::load_asc()
#> ℹ [ INFO ] - Processing block: block_1
#> ✔ [ OK ] - Running eyeris::deblink() for block_1
#> ✔ [ OK ] - Running eyeris::detransient() for block_1
#> ✔ [ OK ] - Running eyeris::interpolate() for block_1
#> ✔ [ OK ] - Running eyeris::lpfilt() for block_1
 #> ! [ SKIP ] - Skipping eyeris::downsample() for block_1
#> ! [ SKIP ] - Skipping eyeris::bin() for block_1
#> ! [ SKIP ] - Skipping eyeris::detrend() for block_1
#> ✔ [ OK ] - Running eyeris::zscore() for block_1
#>
#> Block processing summary:
#> block_1: OK (steps: 6, latest: pupil_raw_deblink_detransient_interpolate_lpfilt_z)
#>
#> ✔ [ OK ] - Running eyeris::summarize_confounds()
#> ! [ INFO ] - Plotting block 1 from possible blocks: 1
#> ℹ [ INFO ] - Plotting with sampling rate: 1000 Hz
#> ! [ SKIP ] - Skipping eyeris::downsample() for block_1
#> ! [ SKIP ] - Skipping eyeris::bin() for block_1
#> ! [ SKIP ] - Skipping eyeris::detrend() for block_1
#> ✔ [ OK ] - Running eyeris::zscore() for block_1
#>
#> Block processing summary:
#> block_1: OK (steps: 6, latest: pupil_raw_deblink_detransient_interpolate_lpfilt_z)
#>
#> ✔ [ OK ] - Running eyeris::summarize_confounds()
#> ! [ INFO ] - Plotting block 1 from possible blocks: 1
#> ℹ [ INFO ] - Plotting with sampling rate: 1000 Hz






