This method provides a structured way to save out pupil data in a BIDS-like structure. The method saves out epoched data as well as the raw pupil timeseries, and formats the directory and filename structures based on the metadata you provide.
Usage
bidsify(
eyeris,
save_all = TRUE,
epochs_list = NULL,
merge_epochs = FALSE,
bids_dir = NULL,
participant_id = NULL,
session_num = NULL,
task_name = NULL,
run_num = NULL,
merge_runs = FALSE,
save_raw = TRUE,
html_report = TRUE,
report_seed = 0,
report_epoch_grouping_var_col = "matched_event",
verbose = TRUE,
pdf_report = deprecated()
)Arguments
- eyeris
An object of class
eyerisderived fromload_asc()- save_all
Logical flag indicating whether all epochs are to be saved or only a subset of them. Defaults to
TRUE- epochs_list
List of epochs to be saved. Defaults to
NULL- merge_epochs
Logical flag indicating whether epochs should be saved as one file or as separate files. Defaults to
FALSE(no merge)- bids_dir
Base bids_directory. Defaults to
NULL- participant_id
BIDS subject ID. Defaults to
NULL- session_num
BIDS session ID. Defaults to
NULL- task_name
BIDS task ID. Defaults to
NULL- run_num
BIDS run ID. Optional override for the run number when there's only one block of data present in a given
.ascfile. This allows you to manually specify a run number (e.g., "03") instead of using the default block number in.ascfiles (1). This is especially useful if you have a single.ascfile for a single run of a task and want your BIDSified derivatives to be labeled correctly. However, for files with multiple recording blocks embedded within the same.ascfile, this parameter is ignored and blocks are automatically numbered as runs (block 1 = run-01, block 2 = run-02, etc.) in the order they appeared/were recorded. Defaults toNULL(no override)- merge_runs
Logical flag indicating whether multiple runs (either from multiple recording blocks existing within the same
.ascfile (see above), or manually specified) should be combined into a single output file. WhenTRUE, adds a 'run' column to identify the source run Defaults toFALSE(i.e., separate files per block/run – the standard BIDS-like-behavior)- save_raw
Logical flag indicating whether to save_raw pupil data in addition to epoched data. Defaults to
TRUE- html_report
Logical flag indicating whether to save out the
eyerispreprocessing summary report as an HTML file. Defaults toTRUE- report_seed
Random seed for the plots that will appear in the report Defaults to
0. Seeplot()for a more detailed description- report_epoch_grouping_var_col
String name of grouping column to use for epoch-by-epoch diagnostic plots in an interactive rendered HTML report. Column name must exist (i.e., be a custom grouping variable name set within the metadata template of your
epoch()call). Defaults to"matched_event", which all epoched dataframes have as a valid column name. To disable these epoch-level diagnostic plots, set toNULL- verbose
A flag to indicate whether to print detailed logging messages. Defaults to
TRUE. Set toFALSEto suppress messages about the current processing step and run silently- pdf_report
(Deprecated) Use
html_report = TRUEinstead
Details
In the future, we intend for this function to save out the data in an official BIDS format for eyetracking data (see the proposal currently under review here). At this time, however, this function instead takes a more BIDS-inspired approach to organizing the output files for preprocessed pupil data.
Examples
# Bleed around blink periods just long enough to remove majority of
# deflections due to eyelid movements
# \donttest{
demo_data <- eyelink_asc_demo_dataset()
# example with unepoched data
demo_data |>
eyeris::glassbox() |>
eyeris::bidsify(
bids_dir = tempdir(), # <- MAKE SURE TO UPDATE TO YOUR DESIRED LOCAL PATH
participant_id = "001",
session_num = "01",
task_name = "assocret",
run_num = "01",
save_raw = TRUE, # save out raw timeseries
html_report = TRUE, # generate interactive report document
report_seed = 0 # make randomly selected plot epochs reproducible
)
#> ✔ [ OK ] - Running eyeris::load_asc()
#> ℹ [ INFO ] - Processing block: block_1
#> ✔ [ OK ] - Running eyeris::deblink() for block_1
#> ✔ [ OK ] - Running eyeris::detransient() for block_1
#> ✔ [ OK ] - Running eyeris::interpolate() for block_1
#> ✔ [ OK ] - Running eyeris::lpfilt() for block_1
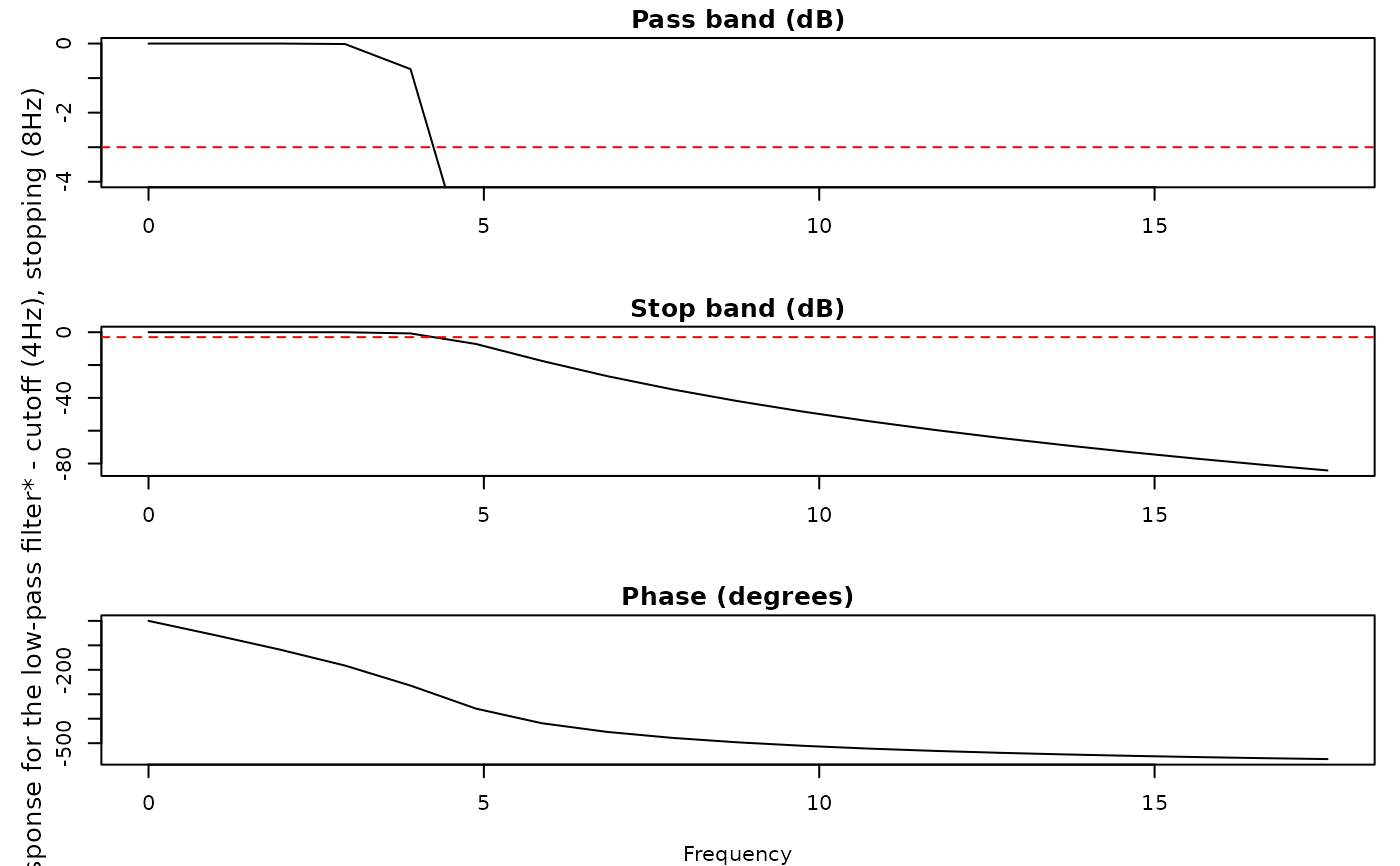 #> ! [ SKIP ] - Skipping eyeris::downsample() for block_1
#> ! [ SKIP ] - Skipping eyeris::bin() for block_1
#> ! [ SKIP ] - Skipping eyeris::detrend() for block_1
#> ✔ [ OK ] - Running eyeris::zscore() for block_1
#>
#> Block processing summary:
#> block_1: OK (steps: 6, latest: pupil_raw_deblink_detransient_interpolate_lpfilt_z)
#>
#> ✔ [ OK ] - Running eyeris::summarize_confounds()
#> ℹ Only 1 block detected...
#> ℹ Using run_num = 01 for single block data
#> ℹ Filtered epochs:
#> ! '/tmp/RtmpjCWoFa' already exists. Skipping creation...
#> ℹ '/tmp/RtmpjCWoFa/derivatives' does not exist. Creating...
#> ✔ BIDS directory successfully created at: '/tmp/RtmpjCWoFa/derivatives'
#> ℹ '/tmp/RtmpjCWoFa/derivatives/sub-001' does not exist. Creating...
#> ✔ BIDS directory successfully created at: '/tmp/RtmpjCWoFa/derivatives/sub-001'
#> ℹ '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01' does not exist. Creating...
#> ✔ BIDS directory successfully created at: '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01'
#> ℹ '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye' does not exist. Creating...
#> ✔ BIDS directory successfully created at: '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye'
#> ℹ Writing single raw pupil timeseries to '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/sub-001_ses-01_task-assocret_run-01_desc-timeseries_pupil.csv'...
#> ✔ Single raw pupil timeseries written to: '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/sub-001_ses-01_task-assocret_run-01_desc-timeseries_pupil.csv'
#> ✔ Confounds file written to: '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/sub-001_ses-01_task-assocret_run-01_desc-confounds.csv'
#> ℹ '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/source/figures' does not exist. Creating...
#> ✔ BIDS directory successfully created at: '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/source/figures'
#> ℹ '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/source/figures/run-01' does not exist. Creating...
#> ✔ BIDS directory successfully created at: '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/source/figures/run-01'
#> ! [ INFO ] - Plotting block 1 from possible blocks: 1
#> ℹ [ INFO ] - Plotting with sampling rate: 1000 Hz
#> ! [ INFO ] - Plotting block 1 from possible blocks: 1
#> ℹ [ INFO ] - Plotting with sampling rate: 1000 Hz
#> ! [ INFO ] - Plotting block 1 from possible blocks: 1
#> ℹ [ INFO ] - Plotting with sampling rate: 1000 Hz
#> ! [ INFO ] - Plotting block 1 from possible blocks: 1
#> ℹ [ INFO ] - Plotting with sampling rate: 1000 Hz
#> ! [ INFO ] - Plotting block 1 from possible blocks: 1
#> ℹ [ INFO ] - Plotting with sampling rate: 1000 Hz
#> ! [ INFO ] - Plotting block 1 from possible blocks: 1
#> ℹ [ INFO ] - Plotting with sampling rate: 1000 Hz
#> ! [ INFO ] - Plotting block 1 from possible blocks: 1
#> ℹ [ INFO ] - Plotting with sampling rate: 1000 Hz
#> ! [ INFO ] - Plotting block 1 from possible blocks: 1
#> ℹ [ INFO ] - Plotting with sampling rate: 1000 Hz
#> ! [ INFO ] - Plotting block 1 from possible blocks: 1
#> ℹ [ INFO ] - Plotting with sampling rate: 1000 Hz
#> ! [ INFO ] - Plotting block 1 from possible blocks: 1
#> ℹ [ INFO ] - Plotting with sampling rate: 1000 Hz
#> ! [ INFO ] - Plotting block 1 from possible blocks: 1
#> ℹ [ INFO ] - Plotting with sampling rate: 1000 Hz
#> ! [ INFO ] - Plotting block 1 from possible blocks: 1
#> ℹ [ INFO ] - Plotting with sampling rate: 1000 Hz
#> ! [ INFO ] - Plotting block 1 from possible blocks: 1
#> ℹ [ INFO ] - Plotting with sampling rate: 1000 Hz
#> ! [ INFO ] - Plotting block 1 from possible blocks: 1
#> ℹ [ INFO ] - Plotting with sampling rate: 1000 Hz
#> ! [ INFO ] - Plotting block 1 from possible blocks: 1
#> ℹ [ INFO ] - Plotting with sampling rate: 1000 Hz
#> ! [ INFO ] - Plotting block 1 from possible blocks: 1
#> ℹ [ INFO ] - Plotting with sampling rate: 1000 Hz
#> ! [ INFO ] - Plotting block 1 from possible blocks: 1
#> ℹ [ INFO ] - Plotting with sampling rate: 1000 Hz
#> ! [ INFO ] - Plotting block 1 from possible blocks: 1
#> ℹ [ INFO ] - Plotting with sampling rate: 1000 Hz
#> ! '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/source/figures/run-01' already exists. Skipping creation...
#> ℹ Created gaze heatmap for run-01
#> [Skipped] No detrend data found for run-01
#>
#>
#> processing file: sub-001.Rmd
#> 1/3
#> 2/3 [citation]
#> 3/3
#> output file: sub-001.knit.md
#> /opt/hostedtoolcache/pandoc/3.1.11/x64/pandoc +RTS -K512m -RTS sub-001.knit.md --to html4 --from markdown+autolink_bare_uris+tex_math_single_backslash --output sub-001.html --lua-filter /home/runner/work/_temp/Library/rmarkdown/rmarkdown/lua/pagebreak.lua --lua-filter /home/runner/work/_temp/Library/rmarkdown/rmarkdown/lua/latex-div.lua --embed-resources --standalone --variable bs3=TRUE --section-divs --table-of-contents --toc-depth 3 --variable toc_float=1 --variable toc_selectors=h1,h2,h3 --variable toc_collapsed=1 --variable toc_smooth_scroll=1 --variable toc_print=1 --template /home/runner/work/_temp/Library/rmarkdown/rmd/h/default.html --no-highlight --variable highlightjs=1 --variable theme=bootstrap --css /home/runner/work/_temp/Library/eyeris/rmarkdown/css/report.css --mathjax --variable 'mathjax-url=https://mathjax.rstudio.com/latest/MathJax.js?config=TeX-AMS-MML_HTMLorMML' --include-in-header /tmp/RtmpjCWoFa/rmarkdown-str1cbd4872a1a5.html
#>
#> Output created: sub-001.html
# example with epoched data
demo_data |>
eyeris::glassbox() |>
eyeris::epoch(
events = "PROBE_{type}_{trial}",
limits = c(-1, 1), # grab 1 second prior to and 1 second post event
label = "prePostProbe" # custom epoch label name
) |>
eyeris::bidsify(
bids_dir = tempdir(), # <- MAKE SURE TO UPDATE TO YOUR DESIRED LOCAL PATH
participant_id = "001",
session_num = "01",
task_name = "assocret",
run_num = "01"
)
#> ✔ [ OK ] - Running eyeris::load_asc()
#> ℹ [ INFO ] - Processing block: block_1
#> ✔ [ OK ] - Running eyeris::deblink() for block_1
#> ✔ [ OK ] - Running eyeris::detransient() for block_1
#> ✔ [ OK ] - Running eyeris::interpolate() for block_1
#> ✔ [ OK ] - Running eyeris::lpfilt() for block_1
#> ! [ SKIP ] - Skipping eyeris::downsample() for block_1
#> ! [ SKIP ] - Skipping eyeris::bin() for block_1
#> ! [ SKIP ] - Skipping eyeris::detrend() for block_1
#> ✔ [ OK ] - Running eyeris::zscore() for block_1
#>
#> Block processing summary:
#> block_1: OK (steps: 6, latest: pupil_raw_deblink_detransient_interpolate_lpfilt_z)
#>
#> ✔ [ OK ] - Running eyeris::summarize_confounds()
#> ℹ Epoching pupil data...
#> ℹ Block 1: found 10 matching events for PROBEtypetrial
#> ✔ Done!
#> ✔ Block 1: pupil data from 10 unique event messages extracted
#> ✔ Pupil epoching completed in 0.21 seconds
#> ℹ Recalculating epoched confounds for new epochs...
#> ℹ Only 1 block detected...
#> ℹ Using run_num = 01 for single block data
#> ℹ Filtered epochs: epoch_prePostProbe
#> ℹ Epochs to save structure:
#> ℹ Names: epoch_prePostProbe
#> ℹ epoch_prePostProbe: list with 2 elements
#> ℹ block_1: data.frame with 20000 rows
#> ℹ info: list
#> ! '/tmp/RtmpjCWoFa' already exists. Skipping creation...
#> ! '/tmp/RtmpjCWoFa/derivatives' already exists. Skipping creation...
#> ! '/tmp/RtmpjCWoFa/derivatives/sub-001' already exists. Skipping creation...
#> ! '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01' already exists. Skipping creation...
#> ! '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye' already exists. Skipping creation...
#> ℹ Processing single-run epoch: epoch_prePostProbe (label: prePostProbe)
#> ℹ Block block_1 for epoch epoch_prePostProbe has 20000 rows
#> No baseline structure found for epoch label: prePostProbe
#> Found epoch events in epoch structure: PROBE_{type}_{trial}
#> No baseline structure found for epoch label: prePostProbe
#> ℹ Writing epoched data for epoch 'prePostProbe' (block block_1) to '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/sub-001_ses-01_task-assocret_run-01_desc-preproc_pupil_epoch-prepostprobe.csv'...
#> ✔ Epoched data for epoch 'prePostProbe' (block block_1) written to: '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/sub-001_ses-01_task-assocret_run-01_desc-preproc_pupil_epoch-prepostprobe.csv'
#> ℹ Writing single raw pupil timeseries to '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/sub-001_ses-01_task-assocret_run-01_desc-timeseries_pupil.csv'...
#> ✔ Single raw pupil timeseries written to: '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/sub-001_ses-01_task-assocret_run-01_desc-timeseries_pupil.csv'
#> ✔ Confounds file written to: '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/sub-001_ses-01_task-assocret_run-01_desc-confounds.csv'
#> No baseline structure found for epoch label: prePostProbe
#> Processing epoch: epoch_prePostProbe -> label: prePostProbe -> baseline:
#> Found epoch events in epoch structure: PROBE_{type}_{trial}
#> No baseline structure found for epoch label: prePostProbe
#> No baseline structure found for epoch label: prePostProbe
#> No baseline structure found for epoch label: prePostProbe
#> No baseline events found for: epoch_prePostProbe
#> No baseline structure found for epoch label: prePostProbe
#> No baseline structure found for epoch label: prePostProbe
#> ℹ Writing epoch summary for block 1 to '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/sub-001_ses-01_task-assocret_run-01_desc-epoch_summary.csv'...
#> ✔ Epoch summary for block 1 written to: '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/sub-001_ses-01_task-assocret_run-01_desc-epoch_summary.csv'
#> No baseline structure found for epoch label: prePostProbe
#> Found epoch events in epoch structure: PROBE_{type}_{trial}
#> No baseline structure found for epoch label: prePostProbe
#> No baseline structure found for epoch label: prePostProbe
#> ℹ Writing epoch-wide confounds for event 'PROBE_START_22'(unique: 'PROBE_START_22') to '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-01_desc-confounds_epoch_wide_PROBE_START_22_epoch-prepostprobe.csv'...
#> ✔ Epoch-wide confounds for event 'PROBE_START_22'(unique: 'PROBE_START_22') written to: '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-01_desc-confounds_epoch_wide_PROBE_START_22_epoch-prepostprobe.csv'
#> ℹ Writing epoch-wide confounds for event 'PROBE_STOP_22'(unique: 'PROBE_STOP_22') to '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-01_desc-confounds_epoch_wide_PROBE_STOP_22_epoch-prepostprobe.csv'...
#> ✔ Epoch-wide confounds for event 'PROBE_STOP_22'(unique: 'PROBE_STOP_22') written to: '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-01_desc-confounds_epoch_wide_PROBE_STOP_22_epoch-prepostprobe.csv'
#> ℹ Writing epoch-wide confounds for event 'PROBE_START_23'(unique: 'PROBE_START_23') to '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-01_desc-confounds_epoch_wide_PROBE_START_23_epoch-prepostprobe.csv'...
#> ✔ Epoch-wide confounds for event 'PROBE_START_23'(unique: 'PROBE_START_23') written to: '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-01_desc-confounds_epoch_wide_PROBE_START_23_epoch-prepostprobe.csv'
#> ℹ Writing epoch-wide confounds for event 'PROBE_STOP_23'(unique: 'PROBE_STOP_23') to '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-01_desc-confounds_epoch_wide_PROBE_STOP_23_epoch-prepostprobe.csv'...
#> ✔ Epoch-wide confounds for event 'PROBE_STOP_23'(unique: 'PROBE_STOP_23') written to: '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-01_desc-confounds_epoch_wide_PROBE_STOP_23_epoch-prepostprobe.csv'
#> ℹ Writing epoch-wide confounds for event 'PROBE_START_24'(unique: 'PROBE_START_24') to '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-01_desc-confounds_epoch_wide_PROBE_START_24_epoch-prepostprobe.csv'...
#> ✔ Epoch-wide confounds for event 'PROBE_START_24'(unique: 'PROBE_START_24') written to: '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-01_desc-confounds_epoch_wide_PROBE_START_24_epoch-prepostprobe.csv'
#> ℹ Writing epoch-wide confounds for event 'PROBE_STOP_24'(unique: 'PROBE_STOP_24') to '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-01_desc-confounds_epoch_wide_PROBE_STOP_24_epoch-prepostprobe.csv'...
#> ✔ Epoch-wide confounds for event 'PROBE_STOP_24'(unique: 'PROBE_STOP_24') written to: '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-01_desc-confounds_epoch_wide_PROBE_STOP_24_epoch-prepostprobe.csv'
#> ℹ Writing epoch-wide confounds for event 'PROBE_START_25'(unique: 'PROBE_START_25') to '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-01_desc-confounds_epoch_wide_PROBE_START_25_epoch-prepostprobe.csv'...
#> ✔ Epoch-wide confounds for event 'PROBE_START_25'(unique: 'PROBE_START_25') written to: '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-01_desc-confounds_epoch_wide_PROBE_START_25_epoch-prepostprobe.csv'
#> ℹ Writing epoch-wide confounds for event 'PROBE_STOP_25'(unique: 'PROBE_STOP_25') to '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-01_desc-confounds_epoch_wide_PROBE_STOP_25_epoch-prepostprobe.csv'...
#> ✔ Epoch-wide confounds for event 'PROBE_STOP_25'(unique: 'PROBE_STOP_25') written to: '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-01_desc-confounds_epoch_wide_PROBE_STOP_25_epoch-prepostprobe.csv'
#> ℹ Writing epoch-wide confounds for event 'PROBE_START_26'(unique: 'PROBE_START_26') to '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-01_desc-confounds_epoch_wide_PROBE_START_26_epoch-prepostprobe.csv'...
#> ✔ Epoch-wide confounds for event 'PROBE_START_26'(unique: 'PROBE_START_26') written to: '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-01_desc-confounds_epoch_wide_PROBE_START_26_epoch-prepostprobe.csv'
#> ℹ Writing epoch-wide confounds for event 'PROBE_STOP_26'(unique: 'PROBE_STOP_26') to '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-01_desc-confounds_epoch_wide_PROBE_STOP_26_epoch-prepostprobe.csv'...
#> ✔ Epoch-wide confounds for event 'PROBE_STOP_26'(unique: 'PROBE_STOP_26') written to: '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-01_desc-confounds_epoch_wide_PROBE_STOP_26_epoch-prepostprobe.csv'
#> No baseline structure found for epoch label: prePostProbe
#> Found epoch events in epoch structure: PROBE_{type}_{trial}
#> No baseline structure found for epoch label: prePostProbe
#> No baseline structure found for epoch label: prePostProbe
#> ℹ Writing step-specific confounds for event 'PROBE_START_22'(unique: 'PROBE_START_22') to '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-01_desc-confounds_steps_PROBE_START_22_epoch-prepostprobe.csv'...
#> ✔ Step-specific confounds for event 'PROBE_START_22'(unique: 'PROBE_START_22') written to: '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-01_desc-confounds_steps_PROBE_START_22_epoch-prepostprobe.csv'
#> ℹ Writing step-specific confounds for event 'PROBE_STOP_22'(unique: 'PROBE_STOP_22') to '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-01_desc-confounds_steps_PROBE_STOP_22_epoch-prepostprobe.csv'...
#> ✔ Step-specific confounds for event 'PROBE_STOP_22'(unique: 'PROBE_STOP_22') written to: '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-01_desc-confounds_steps_PROBE_STOP_22_epoch-prepostprobe.csv'
#> ℹ Writing step-specific confounds for event 'PROBE_START_23'(unique: 'PROBE_START_23') to '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-01_desc-confounds_steps_PROBE_START_23_epoch-prepostprobe.csv'...
#> ✔ Step-specific confounds for event 'PROBE_START_23'(unique: 'PROBE_START_23') written to: '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-01_desc-confounds_steps_PROBE_START_23_epoch-prepostprobe.csv'
#> ℹ Writing step-specific confounds for event 'PROBE_STOP_23'(unique: 'PROBE_STOP_23') to '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-01_desc-confounds_steps_PROBE_STOP_23_epoch-prepostprobe.csv'...
#> ✔ Step-specific confounds for event 'PROBE_STOP_23'(unique: 'PROBE_STOP_23') written to: '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-01_desc-confounds_steps_PROBE_STOP_23_epoch-prepostprobe.csv'
#> ℹ Writing step-specific confounds for event 'PROBE_START_24'(unique: 'PROBE_START_24') to '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-01_desc-confounds_steps_PROBE_START_24_epoch-prepostprobe.csv'...
#> ✔ Step-specific confounds for event 'PROBE_START_24'(unique: 'PROBE_START_24') written to: '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-01_desc-confounds_steps_PROBE_START_24_epoch-prepostprobe.csv'
#> ℹ Writing step-specific confounds for event 'PROBE_STOP_24'(unique: 'PROBE_STOP_24') to '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-01_desc-confounds_steps_PROBE_STOP_24_epoch-prepostprobe.csv'...
#> ✔ Step-specific confounds for event 'PROBE_STOP_24'(unique: 'PROBE_STOP_24') written to: '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-01_desc-confounds_steps_PROBE_STOP_24_epoch-prepostprobe.csv'
#> ℹ Writing step-specific confounds for event 'PROBE_START_25'(unique: 'PROBE_START_25') to '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-01_desc-confounds_steps_PROBE_START_25_epoch-prepostprobe.csv'...
#> ✔ Step-specific confounds for event 'PROBE_START_25'(unique: 'PROBE_START_25') written to: '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-01_desc-confounds_steps_PROBE_START_25_epoch-prepostprobe.csv'
#> ℹ Writing step-specific confounds for event 'PROBE_STOP_25'(unique: 'PROBE_STOP_25') to '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-01_desc-confounds_steps_PROBE_STOP_25_epoch-prepostprobe.csv'...
#> ✔ Step-specific confounds for event 'PROBE_STOP_25'(unique: 'PROBE_STOP_25') written to: '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-01_desc-confounds_steps_PROBE_STOP_25_epoch-prepostprobe.csv'
#> ℹ Writing step-specific confounds for event 'PROBE_START_26'(unique: 'PROBE_START_26') to '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-01_desc-confounds_steps_PROBE_START_26_epoch-prepostprobe.csv'...
#> ✔ Step-specific confounds for event 'PROBE_START_26'(unique: 'PROBE_START_26') written to: '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-01_desc-confounds_steps_PROBE_START_26_epoch-prepostprobe.csv'
#> ℹ Writing step-specific confounds for event 'PROBE_STOP_26'(unique: 'PROBE_STOP_26') to '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-01_desc-confounds_steps_PROBE_STOP_26_epoch-prepostprobe.csv'...
#> ✔ Step-specific confounds for event 'PROBE_STOP_26'(unique: 'PROBE_STOP_26') written to: '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-01_desc-confounds_steps_PROBE_STOP_26_epoch-prepostprobe.csv'
#> ! '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/source/figures' already exists. Skipping creation...
#> ! '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/source/figures/run-01' already exists. Skipping creation...
#> ! [ INFO ] - Plotting block 1 from possible blocks: 1
#> ℹ [ INFO ] - Plotting with sampling rate: 1000 Hz
#> ! [ INFO ] - Plotting block 1 from possible blocks: 1
#> ℹ [ INFO ] - Plotting with sampling rate: 1000 Hz
#> ! [ INFO ] - Plotting block 1 from possible blocks: 1
#> ℹ [ INFO ] - Plotting with sampling rate: 1000 Hz
#> ! [ INFO ] - Plotting block 1 from possible blocks: 1
#> ℹ [ INFO ] - Plotting with sampling rate: 1000 Hz
#> ! [ INFO ] - Plotting block 1 from possible blocks: 1
#> ℹ [ INFO ] - Plotting with sampling rate: 1000 Hz
#> ! [ INFO ] - Plotting block 1 from possible blocks: 1
#> ℹ [ INFO ] - Plotting with sampling rate: 1000 Hz
#> ! [ INFO ] - Plotting block 1 from possible blocks: 1
#> ℹ [ INFO ] - Plotting with sampling rate: 1000 Hz
#> ! [ INFO ] - Plotting block 1 from possible blocks: 1
#> ℹ [ INFO ] - Plotting with sampling rate: 1000 Hz
#> ! [ INFO ] - Plotting block 1 from possible blocks: 1
#> ℹ [ INFO ] - Plotting with sampling rate: 1000 Hz
#> ! [ INFO ] - Plotting block 1 from possible blocks: 1
#> ℹ [ INFO ] - Plotting with sampling rate: 1000 Hz
#> ! [ INFO ] - Plotting block 1 from possible blocks: 1
#> ℹ [ INFO ] - Plotting with sampling rate: 1000 Hz
#> ! [ INFO ] - Plotting block 1 from possible blocks: 1
#> ℹ [ INFO ] - Plotting with sampling rate: 1000 Hz
#> ! [ INFO ] - Plotting block 1 from possible blocks: 1
#> ℹ [ INFO ] - Plotting with sampling rate: 1000 Hz
#> ! [ INFO ] - Plotting block 1 from possible blocks: 1
#> ℹ [ INFO ] - Plotting with sampling rate: 1000 Hz
#> ! [ INFO ] - Plotting block 1 from possible blocks: 1
#> ℹ [ INFO ] - Plotting with sampling rate: 1000 Hz
#> ! [ INFO ] - Plotting block 1 from possible blocks: 1
#> ℹ [ INFO ] - Plotting with sampling rate: 1000 Hz
#> ! [ INFO ] - Plotting block 1 from possible blocks: 1
#> ℹ [ INFO ] - Plotting with sampling rate: 1000 Hz
#> ! [ INFO ] - Plotting block 1 from possible blocks: 1
#> ℹ [ INFO ] - Plotting with sampling rate: 1000 Hz
#> ! '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/source/figures/run-01' already exists. Skipping creation...
#> ℹ Created gaze heatmap for run-01
#> ! '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/source/figures/run-01' already exists. Skipping creation...
#> ℹ '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/source/figures/run-01/epoch_prePostProbe' does not exist. Creating...
#> ✔ BIDS directory successfully created at: '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/source/figures/run-01/epoch_prePostProbe'
#> ℹ Created gaze heatmap for epoch PROBE_START_22 (run-01)
#> ℹ Created gaze heatmap for epoch PROBE_STOP_22 (run-01)
#> ℹ Created gaze heatmap for epoch PROBE_START_23 (run-01)
#> ℹ Created gaze heatmap for epoch PROBE_STOP_23 (run-01)
#> ℹ Created gaze heatmap for epoch PROBE_START_24 (run-01)
#> ℹ Created gaze heatmap for epoch PROBE_STOP_24 (run-01)
#> ℹ Created gaze heatmap for epoch PROBE_START_25 (run-01)
#> ℹ Created gaze heatmap for epoch PROBE_STOP_25 (run-01)
#> ℹ Created gaze heatmap for epoch PROBE_START_26 (run-01)
#> ℹ Created gaze heatmap for epoch PROBE_STOP_26 (run-01)
#>
#>
#> processing file: sub-001_epoch-prePostProbe_run-01.Rmd
#> 1/3
#> 2/3 [citation]
#> 3/3
#> output file: sub-001_epoch-prePostProbe_run-01.knit.md
#> /opt/hostedtoolcache/pandoc/3.1.11/x64/pandoc +RTS -K512m -RTS sub-001_epoch-prePostProbe_run-01.knit.md --to html4 --from markdown+autolink_bare_uris+tex_math_single_backslash --output sub-001_epoch-prePostProbe_run-01.html --lua-filter /home/runner/work/_temp/Library/rmarkdown/rmarkdown/lua/pagebreak.lua --lua-filter /home/runner/work/_temp/Library/rmarkdown/rmarkdown/lua/latex-div.lua --embed-resources --standalone --variable bs3=TRUE --section-divs --template /home/runner/work/_temp/Library/rmarkdown/rmd/h/default.html --no-highlight --variable highlightjs=1 --variable theme=bootstrap --css /home/runner/work/_temp/Library/eyeris/rmarkdown/css/report.css --mathjax --variable 'mathjax-url=https://mathjax.rstudio.com/latest/MathJax.js?config=TeX-AMS-MML_HTMLorMML' --include-in-header /tmp/RtmpjCWoFa/rmarkdown-str1cbd6726b7ff.html
#>
#> Output created: sub-001_epoch-prePostProbe_run-01.html
#> ! Skipping block info for epoch 1 - no valid data
#> [Skipped] No detrend data found for run-01
#>
#>
#> processing file: sub-001.Rmd
#> 1/3
#> 2/3 [citation]
#> 3/3
#> output file: sub-001.knit.md
#> /opt/hostedtoolcache/pandoc/3.1.11/x64/pandoc +RTS -K512m -RTS sub-001.knit.md --to html4 --from markdown+autolink_bare_uris+tex_math_single_backslash --output sub-001.html --lua-filter /home/runner/work/_temp/Library/rmarkdown/rmarkdown/lua/pagebreak.lua --lua-filter /home/runner/work/_temp/Library/rmarkdown/rmarkdown/lua/latex-div.lua --embed-resources --standalone --variable bs3=TRUE --section-divs --table-of-contents --toc-depth 3 --variable toc_float=1 --variable toc_selectors=h1,h2,h3 --variable toc_collapsed=1 --variable toc_smooth_scroll=1 --variable toc_print=1 --template /home/runner/work/_temp/Library/rmarkdown/rmd/h/default.html --no-highlight --variable highlightjs=1 --variable theme=bootstrap --css /home/runner/work/_temp/Library/eyeris/rmarkdown/css/report.css --mathjax --variable 'mathjax-url=https://mathjax.rstudio.com/latest/MathJax.js?config=TeX-AMS-MML_HTMLorMML' --include-in-header /tmp/RtmpjCWoFa/rmarkdown-str1cbd5ab75672.html
#>
#> Output created: sub-001.html
# example with run_num for single block data
demo_data <- eyelink_asc_demo_dataset()
demo_data |>
eyeris::glassbox() |>
eyeris::epoch(
events = "PROBE_{type}_{trial}",
limits = c(-1, 1),
label = "prePostProbe"
) |>
eyeris::bidsify(
bids_dir = tempdir(),
participant_id = "001",
session_num = "01",
task_name = "assocret",
run_num = "03" # override default run-01 (block_1) to use run-03 instead
)
#> ✔ [ OK ] - Running eyeris::load_asc()
#> ℹ [ INFO ] - Processing block: block_1
#> ✔ [ OK ] - Running eyeris::deblink() for block_1
#> ✔ [ OK ] - Running eyeris::detransient() for block_1
#> ✔ [ OK ] - Running eyeris::interpolate() for block_1
#> ✔ [ OK ] - Running eyeris::lpfilt() for block_1
#> ! [ SKIP ] - Skipping eyeris::downsample() for block_1
#> ! [ SKIP ] - Skipping eyeris::bin() for block_1
#> ! [ SKIP ] - Skipping eyeris::detrend() for block_1
#> ✔ [ OK ] - Running eyeris::zscore() for block_1
#>
#> Block processing summary:
#> block_1: OK (steps: 6, latest: pupil_raw_deblink_detransient_interpolate_lpfilt_z)
#>
#> ✔ [ OK ] - Running eyeris::summarize_confounds()
#> ℹ Epoching pupil data...
#> ℹ Block 1: found 10 matching events for PROBEtypetrial
#> ✔ Done!
#> ✔ Block 1: pupil data from 10 unique event messages extracted
#> ✔ Pupil epoching completed in 0.11 seconds
#> ℹ Recalculating epoched confounds for new epochs...
#> ℹ Only 1 block detected...
#> ℹ Using run_num = 03 for single block data
#> ℹ Filtered epochs: epoch_prePostProbe
#> ℹ Epochs to save structure:
#> ℹ Names: epoch_prePostProbe
#> ℹ epoch_prePostProbe: list with 2 elements
#> ℹ block_3: data.frame with 20000 rows
#> ℹ info: list
#> ! '/tmp/RtmpjCWoFa' already exists. Skipping creation...
#> ! '/tmp/RtmpjCWoFa/derivatives' already exists. Skipping creation...
#> ! '/tmp/RtmpjCWoFa/derivatives/sub-001' already exists. Skipping creation...
#> ! '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01' already exists. Skipping creation...
#> ! '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye' already exists. Skipping creation...
#> ℹ Processing single-run epoch: epoch_prePostProbe (label: prePostProbe)
#> ℹ Block block_3 for epoch epoch_prePostProbe has 20000 rows
#> No baseline structure found for epoch label: prePostProbe
#> Found epoch events in epoch structure: PROBE_{type}_{trial}
#> No baseline structure found for epoch label: prePostProbe
#> ℹ Writing epoched data for epoch 'prePostProbe' (block block_3) to '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/sub-001_ses-01_task-assocret_run-03_desc-preproc_pupil_epoch-prepostprobe.csv'...
#> ✔ Epoched data for epoch 'prePostProbe' (block block_3) written to: '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/sub-001_ses-01_task-assocret_run-03_desc-preproc_pupil_epoch-prepostprobe.csv'
#> ℹ Writing single raw pupil timeseries to '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/sub-001_ses-01_task-assocret_run-03_desc-timeseries_pupil.csv'...
#> ✔ Single raw pupil timeseries written to: '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/sub-001_ses-01_task-assocret_run-03_desc-timeseries_pupil.csv'
#> ✔ Confounds file written to: '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/sub-001_ses-01_task-assocret_run-03_desc-confounds.csv'
#> No baseline structure found for epoch label: prePostProbe
#> Processing epoch: epoch_prePostProbe -> label: prePostProbe -> baseline:
#> Found epoch events in epoch structure: PROBE_{type}_{trial}
#> No baseline structure found for epoch label: prePostProbe
#> No baseline structure found for epoch label: prePostProbe
#> No baseline structure found for epoch label: prePostProbe
#> No baseline events found for: epoch_prePostProbe
#> No baseline structure found for epoch label: prePostProbe
#> No baseline structure found for epoch label: prePostProbe
#> ℹ Writing epoch summary for block 3 to '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/sub-001_ses-01_task-assocret_run-03_desc-epoch_summary.csv'...
#> ✔ Epoch summary for block 3 written to: '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/sub-001_ses-01_task-assocret_run-03_desc-epoch_summary.csv'
#> No baseline structure found for epoch label: prePostProbe
#> Found epoch events in epoch structure: PROBE_{type}_{trial}
#> No baseline structure found for epoch label: prePostProbe
#> No baseline structure found for epoch label: prePostProbe
#> ℹ Writing epoch-wide confounds for event 'PROBE_START_22'(unique: 'PROBE_START_22') to '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-03_desc-confounds_epoch_wide_PROBE_START_22_epoch-prepostprobe.csv'...
#> ✔ Epoch-wide confounds for event 'PROBE_START_22'(unique: 'PROBE_START_22') written to: '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-03_desc-confounds_epoch_wide_PROBE_START_22_epoch-prepostprobe.csv'
#> ℹ Writing epoch-wide confounds for event 'PROBE_STOP_22'(unique: 'PROBE_STOP_22') to '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-03_desc-confounds_epoch_wide_PROBE_STOP_22_epoch-prepostprobe.csv'...
#> ✔ Epoch-wide confounds for event 'PROBE_STOP_22'(unique: 'PROBE_STOP_22') written to: '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-03_desc-confounds_epoch_wide_PROBE_STOP_22_epoch-prepostprobe.csv'
#> ℹ Writing epoch-wide confounds for event 'PROBE_START_23'(unique: 'PROBE_START_23') to '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-03_desc-confounds_epoch_wide_PROBE_START_23_epoch-prepostprobe.csv'...
#> ✔ Epoch-wide confounds for event 'PROBE_START_23'(unique: 'PROBE_START_23') written to: '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-03_desc-confounds_epoch_wide_PROBE_START_23_epoch-prepostprobe.csv'
#> ℹ Writing epoch-wide confounds for event 'PROBE_STOP_23'(unique: 'PROBE_STOP_23') to '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-03_desc-confounds_epoch_wide_PROBE_STOP_23_epoch-prepostprobe.csv'...
#> ✔ Epoch-wide confounds for event 'PROBE_STOP_23'(unique: 'PROBE_STOP_23') written to: '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-03_desc-confounds_epoch_wide_PROBE_STOP_23_epoch-prepostprobe.csv'
#> ℹ Writing epoch-wide confounds for event 'PROBE_START_24'(unique: 'PROBE_START_24') to '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-03_desc-confounds_epoch_wide_PROBE_START_24_epoch-prepostprobe.csv'...
#> ✔ Epoch-wide confounds for event 'PROBE_START_24'(unique: 'PROBE_START_24') written to: '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-03_desc-confounds_epoch_wide_PROBE_START_24_epoch-prepostprobe.csv'
#> ℹ Writing epoch-wide confounds for event 'PROBE_STOP_24'(unique: 'PROBE_STOP_24') to '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-03_desc-confounds_epoch_wide_PROBE_STOP_24_epoch-prepostprobe.csv'...
#> ✔ Epoch-wide confounds for event 'PROBE_STOP_24'(unique: 'PROBE_STOP_24') written to: '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-03_desc-confounds_epoch_wide_PROBE_STOP_24_epoch-prepostprobe.csv'
#> ℹ Writing epoch-wide confounds for event 'PROBE_START_25'(unique: 'PROBE_START_25') to '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-03_desc-confounds_epoch_wide_PROBE_START_25_epoch-prepostprobe.csv'...
#> ✔ Epoch-wide confounds for event 'PROBE_START_25'(unique: 'PROBE_START_25') written to: '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-03_desc-confounds_epoch_wide_PROBE_START_25_epoch-prepostprobe.csv'
#> ℹ Writing epoch-wide confounds for event 'PROBE_STOP_25'(unique: 'PROBE_STOP_25') to '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-03_desc-confounds_epoch_wide_PROBE_STOP_25_epoch-prepostprobe.csv'...
#> ✔ Epoch-wide confounds for event 'PROBE_STOP_25'(unique: 'PROBE_STOP_25') written to: '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-03_desc-confounds_epoch_wide_PROBE_STOP_25_epoch-prepostprobe.csv'
#> ℹ Writing epoch-wide confounds for event 'PROBE_START_26'(unique: 'PROBE_START_26') to '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-03_desc-confounds_epoch_wide_PROBE_START_26_epoch-prepostprobe.csv'...
#> ✔ Epoch-wide confounds for event 'PROBE_START_26'(unique: 'PROBE_START_26') written to: '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-03_desc-confounds_epoch_wide_PROBE_START_26_epoch-prepostprobe.csv'
#> ℹ Writing epoch-wide confounds for event 'PROBE_STOP_26'(unique: 'PROBE_STOP_26') to '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-03_desc-confounds_epoch_wide_PROBE_STOP_26_epoch-prepostprobe.csv'...
#> ✔ Epoch-wide confounds for event 'PROBE_STOP_26'(unique: 'PROBE_STOP_26') written to: '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-03_desc-confounds_epoch_wide_PROBE_STOP_26_epoch-prepostprobe.csv'
#> No baseline structure found for epoch label: prePostProbe
#> Found epoch events in epoch structure: PROBE_{type}_{trial}
#> No baseline structure found for epoch label: prePostProbe
#> No baseline structure found for epoch label: prePostProbe
#> ℹ Writing step-specific confounds for event 'PROBE_START_22'(unique: 'PROBE_START_22') to '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-03_desc-confounds_steps_PROBE_START_22_epoch-prepostprobe.csv'...
#> ✔ Step-specific confounds for event 'PROBE_START_22'(unique: 'PROBE_START_22') written to: '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-03_desc-confounds_steps_PROBE_START_22_epoch-prepostprobe.csv'
#> ℹ Writing step-specific confounds for event 'PROBE_STOP_22'(unique: 'PROBE_STOP_22') to '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-03_desc-confounds_steps_PROBE_STOP_22_epoch-prepostprobe.csv'...
#> ✔ Step-specific confounds for event 'PROBE_STOP_22'(unique: 'PROBE_STOP_22') written to: '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-03_desc-confounds_steps_PROBE_STOP_22_epoch-prepostprobe.csv'
#> ℹ Writing step-specific confounds for event 'PROBE_START_23'(unique: 'PROBE_START_23') to '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-03_desc-confounds_steps_PROBE_START_23_epoch-prepostprobe.csv'...
#> ✔ Step-specific confounds for event 'PROBE_START_23'(unique: 'PROBE_START_23') written to: '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-03_desc-confounds_steps_PROBE_START_23_epoch-prepostprobe.csv'
#> ℹ Writing step-specific confounds for event 'PROBE_STOP_23'(unique: 'PROBE_STOP_23') to '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-03_desc-confounds_steps_PROBE_STOP_23_epoch-prepostprobe.csv'...
#> ✔ Step-specific confounds for event 'PROBE_STOP_23'(unique: 'PROBE_STOP_23') written to: '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-03_desc-confounds_steps_PROBE_STOP_23_epoch-prepostprobe.csv'
#> ℹ Writing step-specific confounds for event 'PROBE_START_24'(unique: 'PROBE_START_24') to '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-03_desc-confounds_steps_PROBE_START_24_epoch-prepostprobe.csv'...
#> ✔ Step-specific confounds for event 'PROBE_START_24'(unique: 'PROBE_START_24') written to: '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-03_desc-confounds_steps_PROBE_START_24_epoch-prepostprobe.csv'
#> ℹ Writing step-specific confounds for event 'PROBE_STOP_24'(unique: 'PROBE_STOP_24') to '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-03_desc-confounds_steps_PROBE_STOP_24_epoch-prepostprobe.csv'...
#> ✔ Step-specific confounds for event 'PROBE_STOP_24'(unique: 'PROBE_STOP_24') written to: '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-03_desc-confounds_steps_PROBE_STOP_24_epoch-prepostprobe.csv'
#> ℹ Writing step-specific confounds for event 'PROBE_START_25'(unique: 'PROBE_START_25') to '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-03_desc-confounds_steps_PROBE_START_25_epoch-prepostprobe.csv'...
#> ✔ Step-specific confounds for event 'PROBE_START_25'(unique: 'PROBE_START_25') written to: '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-03_desc-confounds_steps_PROBE_START_25_epoch-prepostprobe.csv'
#> ℹ Writing step-specific confounds for event 'PROBE_STOP_25'(unique: 'PROBE_STOP_25') to '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-03_desc-confounds_steps_PROBE_STOP_25_epoch-prepostprobe.csv'...
#> ✔ Step-specific confounds for event 'PROBE_STOP_25'(unique: 'PROBE_STOP_25') written to: '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-03_desc-confounds_steps_PROBE_STOP_25_epoch-prepostprobe.csv'
#> ℹ Writing step-specific confounds for event 'PROBE_START_26'(unique: 'PROBE_START_26') to '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-03_desc-confounds_steps_PROBE_START_26_epoch-prepostprobe.csv'...
#> ✔ Step-specific confounds for event 'PROBE_START_26'(unique: 'PROBE_START_26') written to: '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-03_desc-confounds_steps_PROBE_START_26_epoch-prepostprobe.csv'
#> ℹ Writing step-specific confounds for event 'PROBE_STOP_26'(unique: 'PROBE_STOP_26') to '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-03_desc-confounds_steps_PROBE_STOP_26_epoch-prepostprobe.csv'...
#> ✔ Step-specific confounds for event 'PROBE_STOP_26'(unique: 'PROBE_STOP_26') written to: '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-03_desc-confounds_steps_PROBE_STOP_26_epoch-prepostprobe.csv'
#> ! '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/source/figures' already exists. Skipping creation...
#> ℹ '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/source/figures/run-03' does not exist. Creating...
#> ✔ BIDS directory successfully created at: '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/source/figures/run-03'
#> ! [ INFO ] - Plotting block 3 from possible blocks: 3
#> ℹ [ INFO ] - Plotting with sampling rate: 1000 Hz
#> ! [ INFO ] - Plotting block 3 from possible blocks: 3
#> ℹ [ INFO ] - Plotting with sampling rate: 1000 Hz
#> ! [ INFO ] - Plotting block 3 from possible blocks: 3
#> ℹ [ INFO ] - Plotting with sampling rate: 1000 Hz
#> ! [ INFO ] - Plotting block 3 from possible blocks: 3
#> ℹ [ INFO ] - Plotting with sampling rate: 1000 Hz
#> ! [ INFO ] - Plotting block 3 from possible blocks: 3
#> ℹ [ INFO ] - Plotting with sampling rate: 1000 Hz
#> ! [ INFO ] - Plotting block 3 from possible blocks: 3
#> ℹ [ INFO ] - Plotting with sampling rate: 1000 Hz
#> ! [ INFO ] - Plotting block 3 from possible blocks: 3
#> ℹ [ INFO ] - Plotting with sampling rate: 1000 Hz
#> ! [ INFO ] - Plotting block 3 from possible blocks: 3
#> ℹ [ INFO ] - Plotting with sampling rate: 1000 Hz
#> ! [ INFO ] - Plotting block 3 from possible blocks: 3
#> ℹ [ INFO ] - Plotting with sampling rate: 1000 Hz
#> ! [ INFO ] - Plotting block 3 from possible blocks: 3
#> ℹ [ INFO ] - Plotting with sampling rate: 1000 Hz
#> ! [ INFO ] - Plotting block 3 from possible blocks: 3
#> ℹ [ INFO ] - Plotting with sampling rate: 1000 Hz
#> ! [ INFO ] - Plotting block 3 from possible blocks: 3
#> ℹ [ INFO ] - Plotting with sampling rate: 1000 Hz
#> ! [ INFO ] - Plotting block 3 from possible blocks: 3
#> ℹ [ INFO ] - Plotting with sampling rate: 1000 Hz
#> ! [ INFO ] - Plotting block 3 from possible blocks: 3
#> ℹ [ INFO ] - Plotting with sampling rate: 1000 Hz
#> ! [ INFO ] - Plotting block 3 from possible blocks: 3
#> ℹ [ INFO ] - Plotting with sampling rate: 1000 Hz
#> ! [ INFO ] - Plotting block 3 from possible blocks: 3
#> ℹ [ INFO ] - Plotting with sampling rate: 1000 Hz
#> ! [ INFO ] - Plotting block 3 from possible blocks: 3
#> ℹ [ INFO ] - Plotting with sampling rate: 1000 Hz
#> ! [ INFO ] - Plotting block 3 from possible blocks: 3
#> ℹ [ INFO ] - Plotting with sampling rate: 1000 Hz
#> ! '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/source/figures/run-03' already exists. Skipping creation...
#> ℹ Created gaze heatmap for run-03
#> ! '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/source/figures/run-03' already exists. Skipping creation...
#> ℹ '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/source/figures/run-03/epoch_prePostProbe' does not exist. Creating...
#> ✔ BIDS directory successfully created at: '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/source/figures/run-03/epoch_prePostProbe'
#> ℹ Created gaze heatmap for epoch PROBE_START_22 (run-03)
#> ℹ Created gaze heatmap for epoch PROBE_STOP_22 (run-03)
#> ℹ Created gaze heatmap for epoch PROBE_START_23 (run-03)
#> ℹ Created gaze heatmap for epoch PROBE_STOP_23 (run-03)
#> ℹ Created gaze heatmap for epoch PROBE_START_24 (run-03)
#> ℹ Created gaze heatmap for epoch PROBE_STOP_24 (run-03)
#> ℹ Created gaze heatmap for epoch PROBE_START_25 (run-03)
#> ℹ Created gaze heatmap for epoch PROBE_STOP_25 (run-03)
#> ℹ Created gaze heatmap for epoch PROBE_START_26 (run-03)
#> ℹ Created gaze heatmap for epoch PROBE_STOP_26 (run-03)
#>
#>
#> processing file: sub-001_epoch-prePostProbe_run-03.Rmd
#> 1/3
#> 2/3 [citation]
#> 3/3
#> output file: sub-001_epoch-prePostProbe_run-03.knit.md
#> /opt/hostedtoolcache/pandoc/3.1.11/x64/pandoc +RTS -K512m -RTS sub-001_epoch-prePostProbe_run-03.knit.md --to html4 --from markdown+autolink_bare_uris+tex_math_single_backslash --output sub-001_epoch-prePostProbe_run-03.html --lua-filter /home/runner/work/_temp/Library/rmarkdown/rmarkdown/lua/pagebreak.lua --lua-filter /home/runner/work/_temp/Library/rmarkdown/rmarkdown/lua/latex-div.lua --embed-resources --standalone --variable bs3=TRUE --section-divs --template /home/runner/work/_temp/Library/rmarkdown/rmd/h/default.html --no-highlight --variable highlightjs=1 --variable theme=bootstrap --css /home/runner/work/_temp/Library/eyeris/rmarkdown/css/report.css --mathjax --variable 'mathjax-url=https://mathjax.rstudio.com/latest/MathJax.js?config=TeX-AMS-MML_HTMLorMML' --include-in-header /tmp/RtmpjCWoFa/rmarkdown-str1cbd3d3462ee.html
#>
#> Output created: sub-001_epoch-prePostProbe_run-03.html
#> ! Skipping block info for epoch 1 - no valid data
#> [Skipped] No detrend data found for run-03
#>
#>
#> processing file: sub-001.Rmd
#> 1/3
#> 2/3 [citation]
#> 3/3
#> output file: sub-001.knit.md
#> /opt/hostedtoolcache/pandoc/3.1.11/x64/pandoc +RTS -K512m -RTS sub-001.knit.md --to html4 --from markdown+autolink_bare_uris+tex_math_single_backslash --output sub-001.html --lua-filter /home/runner/work/_temp/Library/rmarkdown/rmarkdown/lua/pagebreak.lua --lua-filter /home/runner/work/_temp/Library/rmarkdown/rmarkdown/lua/latex-div.lua --embed-resources --standalone --variable bs3=TRUE --section-divs --table-of-contents --toc-depth 3 --variable toc_float=1 --variable toc_selectors=h1,h2,h3 --variable toc_collapsed=1 --variable toc_smooth_scroll=1 --variable toc_print=1 --template /home/runner/work/_temp/Library/rmarkdown/rmd/h/default.html --no-highlight --variable highlightjs=1 --variable theme=bootstrap --css /home/runner/work/_temp/Library/eyeris/rmarkdown/css/report.css --mathjax --variable 'mathjax-url=https://mathjax.rstudio.com/latest/MathJax.js?config=TeX-AMS-MML_HTMLorMML' --include-in-header /tmp/RtmpjCWoFa/rmarkdown-str1cbd52c00ee.html
#>
#> Output created: sub-001.html
# }
#> ! [ SKIP ] - Skipping eyeris::downsample() for block_1
#> ! [ SKIP ] - Skipping eyeris::bin() for block_1
#> ! [ SKIP ] - Skipping eyeris::detrend() for block_1
#> ✔ [ OK ] - Running eyeris::zscore() for block_1
#>
#> Block processing summary:
#> block_1: OK (steps: 6, latest: pupil_raw_deblink_detransient_interpolate_lpfilt_z)
#>
#> ✔ [ OK ] - Running eyeris::summarize_confounds()
#> ℹ Only 1 block detected...
#> ℹ Using run_num = 01 for single block data
#> ℹ Filtered epochs:
#> ! '/tmp/RtmpjCWoFa' already exists. Skipping creation...
#> ℹ '/tmp/RtmpjCWoFa/derivatives' does not exist. Creating...
#> ✔ BIDS directory successfully created at: '/tmp/RtmpjCWoFa/derivatives'
#> ℹ '/tmp/RtmpjCWoFa/derivatives/sub-001' does not exist. Creating...
#> ✔ BIDS directory successfully created at: '/tmp/RtmpjCWoFa/derivatives/sub-001'
#> ℹ '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01' does not exist. Creating...
#> ✔ BIDS directory successfully created at: '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01'
#> ℹ '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye' does not exist. Creating...
#> ✔ BIDS directory successfully created at: '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye'
#> ℹ Writing single raw pupil timeseries to '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/sub-001_ses-01_task-assocret_run-01_desc-timeseries_pupil.csv'...
#> ✔ Single raw pupil timeseries written to: '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/sub-001_ses-01_task-assocret_run-01_desc-timeseries_pupil.csv'
#> ✔ Confounds file written to: '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/sub-001_ses-01_task-assocret_run-01_desc-confounds.csv'
#> ℹ '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/source/figures' does not exist. Creating...
#> ✔ BIDS directory successfully created at: '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/source/figures'
#> ℹ '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/source/figures/run-01' does not exist. Creating...
#> ✔ BIDS directory successfully created at: '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/source/figures/run-01'
#> ! [ INFO ] - Plotting block 1 from possible blocks: 1
#> ℹ [ INFO ] - Plotting with sampling rate: 1000 Hz
#> ! [ INFO ] - Plotting block 1 from possible blocks: 1
#> ℹ [ INFO ] - Plotting with sampling rate: 1000 Hz
#> ! [ INFO ] - Plotting block 1 from possible blocks: 1
#> ℹ [ INFO ] - Plotting with sampling rate: 1000 Hz
#> ! [ INFO ] - Plotting block 1 from possible blocks: 1
#> ℹ [ INFO ] - Plotting with sampling rate: 1000 Hz
#> ! [ INFO ] - Plotting block 1 from possible blocks: 1
#> ℹ [ INFO ] - Plotting with sampling rate: 1000 Hz
#> ! [ INFO ] - Plotting block 1 from possible blocks: 1
#> ℹ [ INFO ] - Plotting with sampling rate: 1000 Hz
#> ! [ INFO ] - Plotting block 1 from possible blocks: 1
#> ℹ [ INFO ] - Plotting with sampling rate: 1000 Hz
#> ! [ INFO ] - Plotting block 1 from possible blocks: 1
#> ℹ [ INFO ] - Plotting with sampling rate: 1000 Hz
#> ! [ INFO ] - Plotting block 1 from possible blocks: 1
#> ℹ [ INFO ] - Plotting with sampling rate: 1000 Hz
#> ! [ INFO ] - Plotting block 1 from possible blocks: 1
#> ℹ [ INFO ] - Plotting with sampling rate: 1000 Hz
#> ! [ INFO ] - Plotting block 1 from possible blocks: 1
#> ℹ [ INFO ] - Plotting with sampling rate: 1000 Hz
#> ! [ INFO ] - Plotting block 1 from possible blocks: 1
#> ℹ [ INFO ] - Plotting with sampling rate: 1000 Hz
#> ! [ INFO ] - Plotting block 1 from possible blocks: 1
#> ℹ [ INFO ] - Plotting with sampling rate: 1000 Hz
#> ! [ INFO ] - Plotting block 1 from possible blocks: 1
#> ℹ [ INFO ] - Plotting with sampling rate: 1000 Hz
#> ! [ INFO ] - Plotting block 1 from possible blocks: 1
#> ℹ [ INFO ] - Plotting with sampling rate: 1000 Hz
#> ! [ INFO ] - Plotting block 1 from possible blocks: 1
#> ℹ [ INFO ] - Plotting with sampling rate: 1000 Hz
#> ! [ INFO ] - Plotting block 1 from possible blocks: 1
#> ℹ [ INFO ] - Plotting with sampling rate: 1000 Hz
#> ! [ INFO ] - Plotting block 1 from possible blocks: 1
#> ℹ [ INFO ] - Plotting with sampling rate: 1000 Hz
#> ! '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/source/figures/run-01' already exists. Skipping creation...
#> ℹ Created gaze heatmap for run-01
#> [Skipped] No detrend data found for run-01
#>
#>
#> processing file: sub-001.Rmd
#> 1/3
#> 2/3 [citation]
#> 3/3
#> output file: sub-001.knit.md
#> /opt/hostedtoolcache/pandoc/3.1.11/x64/pandoc +RTS -K512m -RTS sub-001.knit.md --to html4 --from markdown+autolink_bare_uris+tex_math_single_backslash --output sub-001.html --lua-filter /home/runner/work/_temp/Library/rmarkdown/rmarkdown/lua/pagebreak.lua --lua-filter /home/runner/work/_temp/Library/rmarkdown/rmarkdown/lua/latex-div.lua --embed-resources --standalone --variable bs3=TRUE --section-divs --table-of-contents --toc-depth 3 --variable toc_float=1 --variable toc_selectors=h1,h2,h3 --variable toc_collapsed=1 --variable toc_smooth_scroll=1 --variable toc_print=1 --template /home/runner/work/_temp/Library/rmarkdown/rmd/h/default.html --no-highlight --variable highlightjs=1 --variable theme=bootstrap --css /home/runner/work/_temp/Library/eyeris/rmarkdown/css/report.css --mathjax --variable 'mathjax-url=https://mathjax.rstudio.com/latest/MathJax.js?config=TeX-AMS-MML_HTMLorMML' --include-in-header /tmp/RtmpjCWoFa/rmarkdown-str1cbd4872a1a5.html
#>
#> Output created: sub-001.html
# example with epoched data
demo_data |>
eyeris::glassbox() |>
eyeris::epoch(
events = "PROBE_{type}_{trial}",
limits = c(-1, 1), # grab 1 second prior to and 1 second post event
label = "prePostProbe" # custom epoch label name
) |>
eyeris::bidsify(
bids_dir = tempdir(), # <- MAKE SURE TO UPDATE TO YOUR DESIRED LOCAL PATH
participant_id = "001",
session_num = "01",
task_name = "assocret",
run_num = "01"
)
#> ✔ [ OK ] - Running eyeris::load_asc()
#> ℹ [ INFO ] - Processing block: block_1
#> ✔ [ OK ] - Running eyeris::deblink() for block_1
#> ✔ [ OK ] - Running eyeris::detransient() for block_1
#> ✔ [ OK ] - Running eyeris::interpolate() for block_1
#> ✔ [ OK ] - Running eyeris::lpfilt() for block_1
#> ! [ SKIP ] - Skipping eyeris::downsample() for block_1
#> ! [ SKIP ] - Skipping eyeris::bin() for block_1
#> ! [ SKIP ] - Skipping eyeris::detrend() for block_1
#> ✔ [ OK ] - Running eyeris::zscore() for block_1
#>
#> Block processing summary:
#> block_1: OK (steps: 6, latest: pupil_raw_deblink_detransient_interpolate_lpfilt_z)
#>
#> ✔ [ OK ] - Running eyeris::summarize_confounds()
#> ℹ Epoching pupil data...
#> ℹ Block 1: found 10 matching events for PROBEtypetrial
#> ✔ Done!
#> ✔ Block 1: pupil data from 10 unique event messages extracted
#> ✔ Pupil epoching completed in 0.21 seconds
#> ℹ Recalculating epoched confounds for new epochs...
#> ℹ Only 1 block detected...
#> ℹ Using run_num = 01 for single block data
#> ℹ Filtered epochs: epoch_prePostProbe
#> ℹ Epochs to save structure:
#> ℹ Names: epoch_prePostProbe
#> ℹ epoch_prePostProbe: list with 2 elements
#> ℹ block_1: data.frame with 20000 rows
#> ℹ info: list
#> ! '/tmp/RtmpjCWoFa' already exists. Skipping creation...
#> ! '/tmp/RtmpjCWoFa/derivatives' already exists. Skipping creation...
#> ! '/tmp/RtmpjCWoFa/derivatives/sub-001' already exists. Skipping creation...
#> ! '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01' already exists. Skipping creation...
#> ! '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye' already exists. Skipping creation...
#> ℹ Processing single-run epoch: epoch_prePostProbe (label: prePostProbe)
#> ℹ Block block_1 for epoch epoch_prePostProbe has 20000 rows
#> No baseline structure found for epoch label: prePostProbe
#> Found epoch events in epoch structure: PROBE_{type}_{trial}
#> No baseline structure found for epoch label: prePostProbe
#> ℹ Writing epoched data for epoch 'prePostProbe' (block block_1) to '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/sub-001_ses-01_task-assocret_run-01_desc-preproc_pupil_epoch-prepostprobe.csv'...
#> ✔ Epoched data for epoch 'prePostProbe' (block block_1) written to: '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/sub-001_ses-01_task-assocret_run-01_desc-preproc_pupil_epoch-prepostprobe.csv'
#> ℹ Writing single raw pupil timeseries to '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/sub-001_ses-01_task-assocret_run-01_desc-timeseries_pupil.csv'...
#> ✔ Single raw pupil timeseries written to: '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/sub-001_ses-01_task-assocret_run-01_desc-timeseries_pupil.csv'
#> ✔ Confounds file written to: '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/sub-001_ses-01_task-assocret_run-01_desc-confounds.csv'
#> No baseline structure found for epoch label: prePostProbe
#> Processing epoch: epoch_prePostProbe -> label: prePostProbe -> baseline:
#> Found epoch events in epoch structure: PROBE_{type}_{trial}
#> No baseline structure found for epoch label: prePostProbe
#> No baseline structure found for epoch label: prePostProbe
#> No baseline structure found for epoch label: prePostProbe
#> No baseline events found for: epoch_prePostProbe
#> No baseline structure found for epoch label: prePostProbe
#> No baseline structure found for epoch label: prePostProbe
#> ℹ Writing epoch summary for block 1 to '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/sub-001_ses-01_task-assocret_run-01_desc-epoch_summary.csv'...
#> ✔ Epoch summary for block 1 written to: '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/sub-001_ses-01_task-assocret_run-01_desc-epoch_summary.csv'
#> No baseline structure found for epoch label: prePostProbe
#> Found epoch events in epoch structure: PROBE_{type}_{trial}
#> No baseline structure found for epoch label: prePostProbe
#> No baseline structure found for epoch label: prePostProbe
#> ℹ Writing epoch-wide confounds for event 'PROBE_START_22'(unique: 'PROBE_START_22') to '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-01_desc-confounds_epoch_wide_PROBE_START_22_epoch-prepostprobe.csv'...
#> ✔ Epoch-wide confounds for event 'PROBE_START_22'(unique: 'PROBE_START_22') written to: '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-01_desc-confounds_epoch_wide_PROBE_START_22_epoch-prepostprobe.csv'
#> ℹ Writing epoch-wide confounds for event 'PROBE_STOP_22'(unique: 'PROBE_STOP_22') to '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-01_desc-confounds_epoch_wide_PROBE_STOP_22_epoch-prepostprobe.csv'...
#> ✔ Epoch-wide confounds for event 'PROBE_STOP_22'(unique: 'PROBE_STOP_22') written to: '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-01_desc-confounds_epoch_wide_PROBE_STOP_22_epoch-prepostprobe.csv'
#> ℹ Writing epoch-wide confounds for event 'PROBE_START_23'(unique: 'PROBE_START_23') to '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-01_desc-confounds_epoch_wide_PROBE_START_23_epoch-prepostprobe.csv'...
#> ✔ Epoch-wide confounds for event 'PROBE_START_23'(unique: 'PROBE_START_23') written to: '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-01_desc-confounds_epoch_wide_PROBE_START_23_epoch-prepostprobe.csv'
#> ℹ Writing epoch-wide confounds for event 'PROBE_STOP_23'(unique: 'PROBE_STOP_23') to '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-01_desc-confounds_epoch_wide_PROBE_STOP_23_epoch-prepostprobe.csv'...
#> ✔ Epoch-wide confounds for event 'PROBE_STOP_23'(unique: 'PROBE_STOP_23') written to: '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-01_desc-confounds_epoch_wide_PROBE_STOP_23_epoch-prepostprobe.csv'
#> ℹ Writing epoch-wide confounds for event 'PROBE_START_24'(unique: 'PROBE_START_24') to '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-01_desc-confounds_epoch_wide_PROBE_START_24_epoch-prepostprobe.csv'...
#> ✔ Epoch-wide confounds for event 'PROBE_START_24'(unique: 'PROBE_START_24') written to: '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-01_desc-confounds_epoch_wide_PROBE_START_24_epoch-prepostprobe.csv'
#> ℹ Writing epoch-wide confounds for event 'PROBE_STOP_24'(unique: 'PROBE_STOP_24') to '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-01_desc-confounds_epoch_wide_PROBE_STOP_24_epoch-prepostprobe.csv'...
#> ✔ Epoch-wide confounds for event 'PROBE_STOP_24'(unique: 'PROBE_STOP_24') written to: '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-01_desc-confounds_epoch_wide_PROBE_STOP_24_epoch-prepostprobe.csv'
#> ℹ Writing epoch-wide confounds for event 'PROBE_START_25'(unique: 'PROBE_START_25') to '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-01_desc-confounds_epoch_wide_PROBE_START_25_epoch-prepostprobe.csv'...
#> ✔ Epoch-wide confounds for event 'PROBE_START_25'(unique: 'PROBE_START_25') written to: '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-01_desc-confounds_epoch_wide_PROBE_START_25_epoch-prepostprobe.csv'
#> ℹ Writing epoch-wide confounds for event 'PROBE_STOP_25'(unique: 'PROBE_STOP_25') to '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-01_desc-confounds_epoch_wide_PROBE_STOP_25_epoch-prepostprobe.csv'...
#> ✔ Epoch-wide confounds for event 'PROBE_STOP_25'(unique: 'PROBE_STOP_25') written to: '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-01_desc-confounds_epoch_wide_PROBE_STOP_25_epoch-prepostprobe.csv'
#> ℹ Writing epoch-wide confounds for event 'PROBE_START_26'(unique: 'PROBE_START_26') to '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-01_desc-confounds_epoch_wide_PROBE_START_26_epoch-prepostprobe.csv'...
#> ✔ Epoch-wide confounds for event 'PROBE_START_26'(unique: 'PROBE_START_26') written to: '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-01_desc-confounds_epoch_wide_PROBE_START_26_epoch-prepostprobe.csv'
#> ℹ Writing epoch-wide confounds for event 'PROBE_STOP_26'(unique: 'PROBE_STOP_26') to '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-01_desc-confounds_epoch_wide_PROBE_STOP_26_epoch-prepostprobe.csv'...
#> ✔ Epoch-wide confounds for event 'PROBE_STOP_26'(unique: 'PROBE_STOP_26') written to: '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-01_desc-confounds_epoch_wide_PROBE_STOP_26_epoch-prepostprobe.csv'
#> No baseline structure found for epoch label: prePostProbe
#> Found epoch events in epoch structure: PROBE_{type}_{trial}
#> No baseline structure found for epoch label: prePostProbe
#> No baseline structure found for epoch label: prePostProbe
#> ℹ Writing step-specific confounds for event 'PROBE_START_22'(unique: 'PROBE_START_22') to '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-01_desc-confounds_steps_PROBE_START_22_epoch-prepostprobe.csv'...
#> ✔ Step-specific confounds for event 'PROBE_START_22'(unique: 'PROBE_START_22') written to: '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-01_desc-confounds_steps_PROBE_START_22_epoch-prepostprobe.csv'
#> ℹ Writing step-specific confounds for event 'PROBE_STOP_22'(unique: 'PROBE_STOP_22') to '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-01_desc-confounds_steps_PROBE_STOP_22_epoch-prepostprobe.csv'...
#> ✔ Step-specific confounds for event 'PROBE_STOP_22'(unique: 'PROBE_STOP_22') written to: '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-01_desc-confounds_steps_PROBE_STOP_22_epoch-prepostprobe.csv'
#> ℹ Writing step-specific confounds for event 'PROBE_START_23'(unique: 'PROBE_START_23') to '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-01_desc-confounds_steps_PROBE_START_23_epoch-prepostprobe.csv'...
#> ✔ Step-specific confounds for event 'PROBE_START_23'(unique: 'PROBE_START_23') written to: '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-01_desc-confounds_steps_PROBE_START_23_epoch-prepostprobe.csv'
#> ℹ Writing step-specific confounds for event 'PROBE_STOP_23'(unique: 'PROBE_STOP_23') to '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-01_desc-confounds_steps_PROBE_STOP_23_epoch-prepostprobe.csv'...
#> ✔ Step-specific confounds for event 'PROBE_STOP_23'(unique: 'PROBE_STOP_23') written to: '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-01_desc-confounds_steps_PROBE_STOP_23_epoch-prepostprobe.csv'
#> ℹ Writing step-specific confounds for event 'PROBE_START_24'(unique: 'PROBE_START_24') to '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-01_desc-confounds_steps_PROBE_START_24_epoch-prepostprobe.csv'...
#> ✔ Step-specific confounds for event 'PROBE_START_24'(unique: 'PROBE_START_24') written to: '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-01_desc-confounds_steps_PROBE_START_24_epoch-prepostprobe.csv'
#> ℹ Writing step-specific confounds for event 'PROBE_STOP_24'(unique: 'PROBE_STOP_24') to '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-01_desc-confounds_steps_PROBE_STOP_24_epoch-prepostprobe.csv'...
#> ✔ Step-specific confounds for event 'PROBE_STOP_24'(unique: 'PROBE_STOP_24') written to: '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-01_desc-confounds_steps_PROBE_STOP_24_epoch-prepostprobe.csv'
#> ℹ Writing step-specific confounds for event 'PROBE_START_25'(unique: 'PROBE_START_25') to '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-01_desc-confounds_steps_PROBE_START_25_epoch-prepostprobe.csv'...
#> ✔ Step-specific confounds for event 'PROBE_START_25'(unique: 'PROBE_START_25') written to: '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-01_desc-confounds_steps_PROBE_START_25_epoch-prepostprobe.csv'
#> ℹ Writing step-specific confounds for event 'PROBE_STOP_25'(unique: 'PROBE_STOP_25') to '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-01_desc-confounds_steps_PROBE_STOP_25_epoch-prepostprobe.csv'...
#> ✔ Step-specific confounds for event 'PROBE_STOP_25'(unique: 'PROBE_STOP_25') written to: '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-01_desc-confounds_steps_PROBE_STOP_25_epoch-prepostprobe.csv'
#> ℹ Writing step-specific confounds for event 'PROBE_START_26'(unique: 'PROBE_START_26') to '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-01_desc-confounds_steps_PROBE_START_26_epoch-prepostprobe.csv'...
#> ✔ Step-specific confounds for event 'PROBE_START_26'(unique: 'PROBE_START_26') written to: '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-01_desc-confounds_steps_PROBE_START_26_epoch-prepostprobe.csv'
#> ℹ Writing step-specific confounds for event 'PROBE_STOP_26'(unique: 'PROBE_STOP_26') to '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-01_desc-confounds_steps_PROBE_STOP_26_epoch-prepostprobe.csv'...
#> ✔ Step-specific confounds for event 'PROBE_STOP_26'(unique: 'PROBE_STOP_26') written to: '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-01_desc-confounds_steps_PROBE_STOP_26_epoch-prepostprobe.csv'
#> ! '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/source/figures' already exists. Skipping creation...
#> ! '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/source/figures/run-01' already exists. Skipping creation...
#> ! [ INFO ] - Plotting block 1 from possible blocks: 1
#> ℹ [ INFO ] - Plotting with sampling rate: 1000 Hz
#> ! [ INFO ] - Plotting block 1 from possible blocks: 1
#> ℹ [ INFO ] - Plotting with sampling rate: 1000 Hz
#> ! [ INFO ] - Plotting block 1 from possible blocks: 1
#> ℹ [ INFO ] - Plotting with sampling rate: 1000 Hz
#> ! [ INFO ] - Plotting block 1 from possible blocks: 1
#> ℹ [ INFO ] - Plotting with sampling rate: 1000 Hz
#> ! [ INFO ] - Plotting block 1 from possible blocks: 1
#> ℹ [ INFO ] - Plotting with sampling rate: 1000 Hz
#> ! [ INFO ] - Plotting block 1 from possible blocks: 1
#> ℹ [ INFO ] - Plotting with sampling rate: 1000 Hz
#> ! [ INFO ] - Plotting block 1 from possible blocks: 1
#> ℹ [ INFO ] - Plotting with sampling rate: 1000 Hz
#> ! [ INFO ] - Plotting block 1 from possible blocks: 1
#> ℹ [ INFO ] - Plotting with sampling rate: 1000 Hz
#> ! [ INFO ] - Plotting block 1 from possible blocks: 1
#> ℹ [ INFO ] - Plotting with sampling rate: 1000 Hz
#> ! [ INFO ] - Plotting block 1 from possible blocks: 1
#> ℹ [ INFO ] - Plotting with sampling rate: 1000 Hz
#> ! [ INFO ] - Plotting block 1 from possible blocks: 1
#> ℹ [ INFO ] - Plotting with sampling rate: 1000 Hz
#> ! [ INFO ] - Plotting block 1 from possible blocks: 1
#> ℹ [ INFO ] - Plotting with sampling rate: 1000 Hz
#> ! [ INFO ] - Plotting block 1 from possible blocks: 1
#> ℹ [ INFO ] - Plotting with sampling rate: 1000 Hz
#> ! [ INFO ] - Plotting block 1 from possible blocks: 1
#> ℹ [ INFO ] - Plotting with sampling rate: 1000 Hz
#> ! [ INFO ] - Plotting block 1 from possible blocks: 1
#> ℹ [ INFO ] - Plotting with sampling rate: 1000 Hz
#> ! [ INFO ] - Plotting block 1 from possible blocks: 1
#> ℹ [ INFO ] - Plotting with sampling rate: 1000 Hz
#> ! [ INFO ] - Plotting block 1 from possible blocks: 1
#> ℹ [ INFO ] - Plotting with sampling rate: 1000 Hz
#> ! [ INFO ] - Plotting block 1 from possible blocks: 1
#> ℹ [ INFO ] - Plotting with sampling rate: 1000 Hz
#> ! '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/source/figures/run-01' already exists. Skipping creation...
#> ℹ Created gaze heatmap for run-01
#> ! '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/source/figures/run-01' already exists. Skipping creation...
#> ℹ '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/source/figures/run-01/epoch_prePostProbe' does not exist. Creating...
#> ✔ BIDS directory successfully created at: '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/source/figures/run-01/epoch_prePostProbe'
#> ℹ Created gaze heatmap for epoch PROBE_START_22 (run-01)
#> ℹ Created gaze heatmap for epoch PROBE_STOP_22 (run-01)
#> ℹ Created gaze heatmap for epoch PROBE_START_23 (run-01)
#> ℹ Created gaze heatmap for epoch PROBE_STOP_23 (run-01)
#> ℹ Created gaze heatmap for epoch PROBE_START_24 (run-01)
#> ℹ Created gaze heatmap for epoch PROBE_STOP_24 (run-01)
#> ℹ Created gaze heatmap for epoch PROBE_START_25 (run-01)
#> ℹ Created gaze heatmap for epoch PROBE_STOP_25 (run-01)
#> ℹ Created gaze heatmap for epoch PROBE_START_26 (run-01)
#> ℹ Created gaze heatmap for epoch PROBE_STOP_26 (run-01)
#>
#>
#> processing file: sub-001_epoch-prePostProbe_run-01.Rmd
#> 1/3
#> 2/3 [citation]
#> 3/3
#> output file: sub-001_epoch-prePostProbe_run-01.knit.md
#> /opt/hostedtoolcache/pandoc/3.1.11/x64/pandoc +RTS -K512m -RTS sub-001_epoch-prePostProbe_run-01.knit.md --to html4 --from markdown+autolink_bare_uris+tex_math_single_backslash --output sub-001_epoch-prePostProbe_run-01.html --lua-filter /home/runner/work/_temp/Library/rmarkdown/rmarkdown/lua/pagebreak.lua --lua-filter /home/runner/work/_temp/Library/rmarkdown/rmarkdown/lua/latex-div.lua --embed-resources --standalone --variable bs3=TRUE --section-divs --template /home/runner/work/_temp/Library/rmarkdown/rmd/h/default.html --no-highlight --variable highlightjs=1 --variable theme=bootstrap --css /home/runner/work/_temp/Library/eyeris/rmarkdown/css/report.css --mathjax --variable 'mathjax-url=https://mathjax.rstudio.com/latest/MathJax.js?config=TeX-AMS-MML_HTMLorMML' --include-in-header /tmp/RtmpjCWoFa/rmarkdown-str1cbd6726b7ff.html
#>
#> Output created: sub-001_epoch-prePostProbe_run-01.html
#> ! Skipping block info for epoch 1 - no valid data
#> [Skipped] No detrend data found for run-01
#>
#>
#> processing file: sub-001.Rmd
#> 1/3
#> 2/3 [citation]
#> 3/3
#> output file: sub-001.knit.md
#> /opt/hostedtoolcache/pandoc/3.1.11/x64/pandoc +RTS -K512m -RTS sub-001.knit.md --to html4 --from markdown+autolink_bare_uris+tex_math_single_backslash --output sub-001.html --lua-filter /home/runner/work/_temp/Library/rmarkdown/rmarkdown/lua/pagebreak.lua --lua-filter /home/runner/work/_temp/Library/rmarkdown/rmarkdown/lua/latex-div.lua --embed-resources --standalone --variable bs3=TRUE --section-divs --table-of-contents --toc-depth 3 --variable toc_float=1 --variable toc_selectors=h1,h2,h3 --variable toc_collapsed=1 --variable toc_smooth_scroll=1 --variable toc_print=1 --template /home/runner/work/_temp/Library/rmarkdown/rmd/h/default.html --no-highlight --variable highlightjs=1 --variable theme=bootstrap --css /home/runner/work/_temp/Library/eyeris/rmarkdown/css/report.css --mathjax --variable 'mathjax-url=https://mathjax.rstudio.com/latest/MathJax.js?config=TeX-AMS-MML_HTMLorMML' --include-in-header /tmp/RtmpjCWoFa/rmarkdown-str1cbd5ab75672.html
#>
#> Output created: sub-001.html
# example with run_num for single block data
demo_data <- eyelink_asc_demo_dataset()
demo_data |>
eyeris::glassbox() |>
eyeris::epoch(
events = "PROBE_{type}_{trial}",
limits = c(-1, 1),
label = "prePostProbe"
) |>
eyeris::bidsify(
bids_dir = tempdir(),
participant_id = "001",
session_num = "01",
task_name = "assocret",
run_num = "03" # override default run-01 (block_1) to use run-03 instead
)
#> ✔ [ OK ] - Running eyeris::load_asc()
#> ℹ [ INFO ] - Processing block: block_1
#> ✔ [ OK ] - Running eyeris::deblink() for block_1
#> ✔ [ OK ] - Running eyeris::detransient() for block_1
#> ✔ [ OK ] - Running eyeris::interpolate() for block_1
#> ✔ [ OK ] - Running eyeris::lpfilt() for block_1
#> ! [ SKIP ] - Skipping eyeris::downsample() for block_1
#> ! [ SKIP ] - Skipping eyeris::bin() for block_1
#> ! [ SKIP ] - Skipping eyeris::detrend() for block_1
#> ✔ [ OK ] - Running eyeris::zscore() for block_1
#>
#> Block processing summary:
#> block_1: OK (steps: 6, latest: pupil_raw_deblink_detransient_interpolate_lpfilt_z)
#>
#> ✔ [ OK ] - Running eyeris::summarize_confounds()
#> ℹ Epoching pupil data...
#> ℹ Block 1: found 10 matching events for PROBEtypetrial
#> ✔ Done!
#> ✔ Block 1: pupil data from 10 unique event messages extracted
#> ✔ Pupil epoching completed in 0.11 seconds
#> ℹ Recalculating epoched confounds for new epochs...
#> ℹ Only 1 block detected...
#> ℹ Using run_num = 03 for single block data
#> ℹ Filtered epochs: epoch_prePostProbe
#> ℹ Epochs to save structure:
#> ℹ Names: epoch_prePostProbe
#> ℹ epoch_prePostProbe: list with 2 elements
#> ℹ block_3: data.frame with 20000 rows
#> ℹ info: list
#> ! '/tmp/RtmpjCWoFa' already exists. Skipping creation...
#> ! '/tmp/RtmpjCWoFa/derivatives' already exists. Skipping creation...
#> ! '/tmp/RtmpjCWoFa/derivatives/sub-001' already exists. Skipping creation...
#> ! '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01' already exists. Skipping creation...
#> ! '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye' already exists. Skipping creation...
#> ℹ Processing single-run epoch: epoch_prePostProbe (label: prePostProbe)
#> ℹ Block block_3 for epoch epoch_prePostProbe has 20000 rows
#> No baseline structure found for epoch label: prePostProbe
#> Found epoch events in epoch structure: PROBE_{type}_{trial}
#> No baseline structure found for epoch label: prePostProbe
#> ℹ Writing epoched data for epoch 'prePostProbe' (block block_3) to '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/sub-001_ses-01_task-assocret_run-03_desc-preproc_pupil_epoch-prepostprobe.csv'...
#> ✔ Epoched data for epoch 'prePostProbe' (block block_3) written to: '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/sub-001_ses-01_task-assocret_run-03_desc-preproc_pupil_epoch-prepostprobe.csv'
#> ℹ Writing single raw pupil timeseries to '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/sub-001_ses-01_task-assocret_run-03_desc-timeseries_pupil.csv'...
#> ✔ Single raw pupil timeseries written to: '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/sub-001_ses-01_task-assocret_run-03_desc-timeseries_pupil.csv'
#> ✔ Confounds file written to: '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/sub-001_ses-01_task-assocret_run-03_desc-confounds.csv'
#> No baseline structure found for epoch label: prePostProbe
#> Processing epoch: epoch_prePostProbe -> label: prePostProbe -> baseline:
#> Found epoch events in epoch structure: PROBE_{type}_{trial}
#> No baseline structure found for epoch label: prePostProbe
#> No baseline structure found for epoch label: prePostProbe
#> No baseline structure found for epoch label: prePostProbe
#> No baseline events found for: epoch_prePostProbe
#> No baseline structure found for epoch label: prePostProbe
#> No baseline structure found for epoch label: prePostProbe
#> ℹ Writing epoch summary for block 3 to '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/sub-001_ses-01_task-assocret_run-03_desc-epoch_summary.csv'...
#> ✔ Epoch summary for block 3 written to: '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/sub-001_ses-01_task-assocret_run-03_desc-epoch_summary.csv'
#> No baseline structure found for epoch label: prePostProbe
#> Found epoch events in epoch structure: PROBE_{type}_{trial}
#> No baseline structure found for epoch label: prePostProbe
#> No baseline structure found for epoch label: prePostProbe
#> ℹ Writing epoch-wide confounds for event 'PROBE_START_22'(unique: 'PROBE_START_22') to '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-03_desc-confounds_epoch_wide_PROBE_START_22_epoch-prepostprobe.csv'...
#> ✔ Epoch-wide confounds for event 'PROBE_START_22'(unique: 'PROBE_START_22') written to: '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-03_desc-confounds_epoch_wide_PROBE_START_22_epoch-prepostprobe.csv'
#> ℹ Writing epoch-wide confounds for event 'PROBE_STOP_22'(unique: 'PROBE_STOP_22') to '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-03_desc-confounds_epoch_wide_PROBE_STOP_22_epoch-prepostprobe.csv'...
#> ✔ Epoch-wide confounds for event 'PROBE_STOP_22'(unique: 'PROBE_STOP_22') written to: '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-03_desc-confounds_epoch_wide_PROBE_STOP_22_epoch-prepostprobe.csv'
#> ℹ Writing epoch-wide confounds for event 'PROBE_START_23'(unique: 'PROBE_START_23') to '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-03_desc-confounds_epoch_wide_PROBE_START_23_epoch-prepostprobe.csv'...
#> ✔ Epoch-wide confounds for event 'PROBE_START_23'(unique: 'PROBE_START_23') written to: '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-03_desc-confounds_epoch_wide_PROBE_START_23_epoch-prepostprobe.csv'
#> ℹ Writing epoch-wide confounds for event 'PROBE_STOP_23'(unique: 'PROBE_STOP_23') to '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-03_desc-confounds_epoch_wide_PROBE_STOP_23_epoch-prepostprobe.csv'...
#> ✔ Epoch-wide confounds for event 'PROBE_STOP_23'(unique: 'PROBE_STOP_23') written to: '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-03_desc-confounds_epoch_wide_PROBE_STOP_23_epoch-prepostprobe.csv'
#> ℹ Writing epoch-wide confounds for event 'PROBE_START_24'(unique: 'PROBE_START_24') to '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-03_desc-confounds_epoch_wide_PROBE_START_24_epoch-prepostprobe.csv'...
#> ✔ Epoch-wide confounds for event 'PROBE_START_24'(unique: 'PROBE_START_24') written to: '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-03_desc-confounds_epoch_wide_PROBE_START_24_epoch-prepostprobe.csv'
#> ℹ Writing epoch-wide confounds for event 'PROBE_STOP_24'(unique: 'PROBE_STOP_24') to '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-03_desc-confounds_epoch_wide_PROBE_STOP_24_epoch-prepostprobe.csv'...
#> ✔ Epoch-wide confounds for event 'PROBE_STOP_24'(unique: 'PROBE_STOP_24') written to: '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-03_desc-confounds_epoch_wide_PROBE_STOP_24_epoch-prepostprobe.csv'
#> ℹ Writing epoch-wide confounds for event 'PROBE_START_25'(unique: 'PROBE_START_25') to '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-03_desc-confounds_epoch_wide_PROBE_START_25_epoch-prepostprobe.csv'...
#> ✔ Epoch-wide confounds for event 'PROBE_START_25'(unique: 'PROBE_START_25') written to: '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-03_desc-confounds_epoch_wide_PROBE_START_25_epoch-prepostprobe.csv'
#> ℹ Writing epoch-wide confounds for event 'PROBE_STOP_25'(unique: 'PROBE_STOP_25') to '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-03_desc-confounds_epoch_wide_PROBE_STOP_25_epoch-prepostprobe.csv'...
#> ✔ Epoch-wide confounds for event 'PROBE_STOP_25'(unique: 'PROBE_STOP_25') written to: '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-03_desc-confounds_epoch_wide_PROBE_STOP_25_epoch-prepostprobe.csv'
#> ℹ Writing epoch-wide confounds for event 'PROBE_START_26'(unique: 'PROBE_START_26') to '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-03_desc-confounds_epoch_wide_PROBE_START_26_epoch-prepostprobe.csv'...
#> ✔ Epoch-wide confounds for event 'PROBE_START_26'(unique: 'PROBE_START_26') written to: '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-03_desc-confounds_epoch_wide_PROBE_START_26_epoch-prepostprobe.csv'
#> ℹ Writing epoch-wide confounds for event 'PROBE_STOP_26'(unique: 'PROBE_STOP_26') to '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-03_desc-confounds_epoch_wide_PROBE_STOP_26_epoch-prepostprobe.csv'...
#> ✔ Epoch-wide confounds for event 'PROBE_STOP_26'(unique: 'PROBE_STOP_26') written to: '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-03_desc-confounds_epoch_wide_PROBE_STOP_26_epoch-prepostprobe.csv'
#> No baseline structure found for epoch label: prePostProbe
#> Found epoch events in epoch structure: PROBE_{type}_{trial}
#> No baseline structure found for epoch label: prePostProbe
#> No baseline structure found for epoch label: prePostProbe
#> ℹ Writing step-specific confounds for event 'PROBE_START_22'(unique: 'PROBE_START_22') to '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-03_desc-confounds_steps_PROBE_START_22_epoch-prepostprobe.csv'...
#> ✔ Step-specific confounds for event 'PROBE_START_22'(unique: 'PROBE_START_22') written to: '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-03_desc-confounds_steps_PROBE_START_22_epoch-prepostprobe.csv'
#> ℹ Writing step-specific confounds for event 'PROBE_STOP_22'(unique: 'PROBE_STOP_22') to '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-03_desc-confounds_steps_PROBE_STOP_22_epoch-prepostprobe.csv'...
#> ✔ Step-specific confounds for event 'PROBE_STOP_22'(unique: 'PROBE_STOP_22') written to: '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-03_desc-confounds_steps_PROBE_STOP_22_epoch-prepostprobe.csv'
#> ℹ Writing step-specific confounds for event 'PROBE_START_23'(unique: 'PROBE_START_23') to '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-03_desc-confounds_steps_PROBE_START_23_epoch-prepostprobe.csv'...
#> ✔ Step-specific confounds for event 'PROBE_START_23'(unique: 'PROBE_START_23') written to: '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-03_desc-confounds_steps_PROBE_START_23_epoch-prepostprobe.csv'
#> ℹ Writing step-specific confounds for event 'PROBE_STOP_23'(unique: 'PROBE_STOP_23') to '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-03_desc-confounds_steps_PROBE_STOP_23_epoch-prepostprobe.csv'...
#> ✔ Step-specific confounds for event 'PROBE_STOP_23'(unique: 'PROBE_STOP_23') written to: '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-03_desc-confounds_steps_PROBE_STOP_23_epoch-prepostprobe.csv'
#> ℹ Writing step-specific confounds for event 'PROBE_START_24'(unique: 'PROBE_START_24') to '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-03_desc-confounds_steps_PROBE_START_24_epoch-prepostprobe.csv'...
#> ✔ Step-specific confounds for event 'PROBE_START_24'(unique: 'PROBE_START_24') written to: '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-03_desc-confounds_steps_PROBE_START_24_epoch-prepostprobe.csv'
#> ℹ Writing step-specific confounds for event 'PROBE_STOP_24'(unique: 'PROBE_STOP_24') to '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-03_desc-confounds_steps_PROBE_STOP_24_epoch-prepostprobe.csv'...
#> ✔ Step-specific confounds for event 'PROBE_STOP_24'(unique: 'PROBE_STOP_24') written to: '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-03_desc-confounds_steps_PROBE_STOP_24_epoch-prepostprobe.csv'
#> ℹ Writing step-specific confounds for event 'PROBE_START_25'(unique: 'PROBE_START_25') to '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-03_desc-confounds_steps_PROBE_START_25_epoch-prepostprobe.csv'...
#> ✔ Step-specific confounds for event 'PROBE_START_25'(unique: 'PROBE_START_25') written to: '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-03_desc-confounds_steps_PROBE_START_25_epoch-prepostprobe.csv'
#> ℹ Writing step-specific confounds for event 'PROBE_STOP_25'(unique: 'PROBE_STOP_25') to '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-03_desc-confounds_steps_PROBE_STOP_25_epoch-prepostprobe.csv'...
#> ✔ Step-specific confounds for event 'PROBE_STOP_25'(unique: 'PROBE_STOP_25') written to: '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-03_desc-confounds_steps_PROBE_STOP_25_epoch-prepostprobe.csv'
#> ℹ Writing step-specific confounds for event 'PROBE_START_26'(unique: 'PROBE_START_26') to '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-03_desc-confounds_steps_PROBE_START_26_epoch-prepostprobe.csv'...
#> ✔ Step-specific confounds for event 'PROBE_START_26'(unique: 'PROBE_START_26') written to: '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-03_desc-confounds_steps_PROBE_START_26_epoch-prepostprobe.csv'
#> ℹ Writing step-specific confounds for event 'PROBE_STOP_26'(unique: 'PROBE_STOP_26') to '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-03_desc-confounds_steps_PROBE_STOP_26_epoch-prepostprobe.csv'...
#> ✔ Step-specific confounds for event 'PROBE_STOP_26'(unique: 'PROBE_STOP_26') written to: '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/eye/epoch_prePostProbe/sub-001_ses-01_task-assocret_run-03_desc-confounds_steps_PROBE_STOP_26_epoch-prepostprobe.csv'
#> ! '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/source/figures' already exists. Skipping creation...
#> ℹ '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/source/figures/run-03' does not exist. Creating...
#> ✔ BIDS directory successfully created at: '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/source/figures/run-03'
#> ! [ INFO ] - Plotting block 3 from possible blocks: 3
#> ℹ [ INFO ] - Plotting with sampling rate: 1000 Hz
#> ! [ INFO ] - Plotting block 3 from possible blocks: 3
#> ℹ [ INFO ] - Plotting with sampling rate: 1000 Hz
#> ! [ INFO ] - Plotting block 3 from possible blocks: 3
#> ℹ [ INFO ] - Plotting with sampling rate: 1000 Hz
#> ! [ INFO ] - Plotting block 3 from possible blocks: 3
#> ℹ [ INFO ] - Plotting with sampling rate: 1000 Hz
#> ! [ INFO ] - Plotting block 3 from possible blocks: 3
#> ℹ [ INFO ] - Plotting with sampling rate: 1000 Hz
#> ! [ INFO ] - Plotting block 3 from possible blocks: 3
#> ℹ [ INFO ] - Plotting with sampling rate: 1000 Hz
#> ! [ INFO ] - Plotting block 3 from possible blocks: 3
#> ℹ [ INFO ] - Plotting with sampling rate: 1000 Hz
#> ! [ INFO ] - Plotting block 3 from possible blocks: 3
#> ℹ [ INFO ] - Plotting with sampling rate: 1000 Hz
#> ! [ INFO ] - Plotting block 3 from possible blocks: 3
#> ℹ [ INFO ] - Plotting with sampling rate: 1000 Hz
#> ! [ INFO ] - Plotting block 3 from possible blocks: 3
#> ℹ [ INFO ] - Plotting with sampling rate: 1000 Hz
#> ! [ INFO ] - Plotting block 3 from possible blocks: 3
#> ℹ [ INFO ] - Plotting with sampling rate: 1000 Hz
#> ! [ INFO ] - Plotting block 3 from possible blocks: 3
#> ℹ [ INFO ] - Plotting with sampling rate: 1000 Hz
#> ! [ INFO ] - Plotting block 3 from possible blocks: 3
#> ℹ [ INFO ] - Plotting with sampling rate: 1000 Hz
#> ! [ INFO ] - Plotting block 3 from possible blocks: 3
#> ℹ [ INFO ] - Plotting with sampling rate: 1000 Hz
#> ! [ INFO ] - Plotting block 3 from possible blocks: 3
#> ℹ [ INFO ] - Plotting with sampling rate: 1000 Hz
#> ! [ INFO ] - Plotting block 3 from possible blocks: 3
#> ℹ [ INFO ] - Plotting with sampling rate: 1000 Hz
#> ! [ INFO ] - Plotting block 3 from possible blocks: 3
#> ℹ [ INFO ] - Plotting with sampling rate: 1000 Hz
#> ! [ INFO ] - Plotting block 3 from possible blocks: 3
#> ℹ [ INFO ] - Plotting with sampling rate: 1000 Hz
#> ! '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/source/figures/run-03' already exists. Skipping creation...
#> ℹ Created gaze heatmap for run-03
#> ! '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/source/figures/run-03' already exists. Skipping creation...
#> ℹ '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/source/figures/run-03/epoch_prePostProbe' does not exist. Creating...
#> ✔ BIDS directory successfully created at: '/tmp/RtmpjCWoFa/derivatives/sub-001/ses-01/source/figures/run-03/epoch_prePostProbe'
#> ℹ Created gaze heatmap for epoch PROBE_START_22 (run-03)
#> ℹ Created gaze heatmap for epoch PROBE_STOP_22 (run-03)
#> ℹ Created gaze heatmap for epoch PROBE_START_23 (run-03)
#> ℹ Created gaze heatmap for epoch PROBE_STOP_23 (run-03)
#> ℹ Created gaze heatmap for epoch PROBE_START_24 (run-03)
#> ℹ Created gaze heatmap for epoch PROBE_STOP_24 (run-03)
#> ℹ Created gaze heatmap for epoch PROBE_START_25 (run-03)
#> ℹ Created gaze heatmap for epoch PROBE_STOP_25 (run-03)
#> ℹ Created gaze heatmap for epoch PROBE_START_26 (run-03)
#> ℹ Created gaze heatmap for epoch PROBE_STOP_26 (run-03)
#>
#>
#> processing file: sub-001_epoch-prePostProbe_run-03.Rmd
#> 1/3
#> 2/3 [citation]
#> 3/3
#> output file: sub-001_epoch-prePostProbe_run-03.knit.md
#> /opt/hostedtoolcache/pandoc/3.1.11/x64/pandoc +RTS -K512m -RTS sub-001_epoch-prePostProbe_run-03.knit.md --to html4 --from markdown+autolink_bare_uris+tex_math_single_backslash --output sub-001_epoch-prePostProbe_run-03.html --lua-filter /home/runner/work/_temp/Library/rmarkdown/rmarkdown/lua/pagebreak.lua --lua-filter /home/runner/work/_temp/Library/rmarkdown/rmarkdown/lua/latex-div.lua --embed-resources --standalone --variable bs3=TRUE --section-divs --template /home/runner/work/_temp/Library/rmarkdown/rmd/h/default.html --no-highlight --variable highlightjs=1 --variable theme=bootstrap --css /home/runner/work/_temp/Library/eyeris/rmarkdown/css/report.css --mathjax --variable 'mathjax-url=https://mathjax.rstudio.com/latest/MathJax.js?config=TeX-AMS-MML_HTMLorMML' --include-in-header /tmp/RtmpjCWoFa/rmarkdown-str1cbd3d3462ee.html
#>
#> Output created: sub-001_epoch-prePostProbe_run-03.html
#> ! Skipping block info for epoch 1 - no valid data
#> [Skipped] No detrend data found for run-03
#>
#>
#> processing file: sub-001.Rmd
#> 1/3
#> 2/3 [citation]
#> 3/3
#> output file: sub-001.knit.md
#> /opt/hostedtoolcache/pandoc/3.1.11/x64/pandoc +RTS -K512m -RTS sub-001.knit.md --to html4 --from markdown+autolink_bare_uris+tex_math_single_backslash --output sub-001.html --lua-filter /home/runner/work/_temp/Library/rmarkdown/rmarkdown/lua/pagebreak.lua --lua-filter /home/runner/work/_temp/Library/rmarkdown/rmarkdown/lua/latex-div.lua --embed-resources --standalone --variable bs3=TRUE --section-divs --table-of-contents --toc-depth 3 --variable toc_float=1 --variable toc_selectors=h1,h2,h3 --variable toc_collapsed=1 --variable toc_smooth_scroll=1 --variable toc_print=1 --template /home/runner/work/_temp/Library/rmarkdown/rmd/h/default.html --no-highlight --variable highlightjs=1 --variable theme=bootstrap --css /home/runner/work/_temp/Library/eyeris/rmarkdown/css/report.css --mathjax --variable 'mathjax-url=https://mathjax.rstudio.com/latest/MathJax.js?config=TeX-AMS-MML_HTMLorMML' --include-in-header /tmp/RtmpjCWoFa/rmarkdown-str1cbd52c00ee.html
#>
#> Output created: sub-001.html
# }
